1166
Coil Sensitivity Estimation with Deep Sets Towards End-to-End Accelerated MRI Reconstruction1Digital Technology and Innovation, Siemens Healthineers, Princeton, NJ, United States, 2Magnetic Resonance, Siemens Healthineers, Erlangen, Germany
Synopsis
Parallel Imaging (PI) is a crucial technique for accelerating data acquisition in Magnetic Resonance Imaging (MRI), which is exceedingly time-consuming. With current SENSE-based MRI reconstruction formulated as a trainable unrolled optimization framework with several cascades of regularization networks and varying data consistency layers, coils sensitivity maps (CSMs) are needed at each cascade. Therefore, we propose a deep sets CSM estimation network (DS-CSME in short), enabling an end-to-end deep learning solution that allows for further MRI acceleration while preserving the overall reconstructed image quality.
Introduction
Parallel Imaging (PI) is a crucial technique for accelerating data acquisition in Magnetic Resonance Imaging (MRI), which is exceedingly time-consuming. In Parallel Imaging (PI), multiple receiver coils are utilized to simultaneously acquire various views of the underlying anatomy, which are then optimally combined using coils sensitivity maps (CSMs) estimated using the fully sampled central region of k-space corresponding to low frequencies called the Auto-Calibration Signal (ACS). In ESPIRiT [1], for each pixel location, the reconstruction operator's eigendecomposition is applied in the image domain to obtain the required CSMs. Such an approach has several limitations, including computational complexity (time-consuming). Calibrationless methods have been proposed before, but they also suffer from the high cost of solving the resulting nonlinear least-square problem [2, 3]. With the increasing popularity of deep learning (DL) accelerated MRI reconstruction models, an approach for learning the needed CSMs is highly desirable to enable an end-to-end framework for optimal reconstructions. We propose a CSM estimation approach that can handle unordered and varying-in-number k-space multi-coil data.Methods
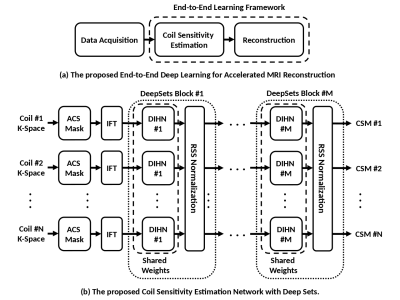
With current SENSE-based MRI reconstruction formulated as a trainable unrolled optimization framework with several cascades of regularization networks and varying data consistency layers, CSMs are needed at each cascade. Therefore, as shown in Figure 1a, we propose a deep sets CSM estimation network (DS-CSME in short), enabling an end-to-end deep learning solution that allows for further MRI acceleration while preserving the overall reconstructed image quality.Figure 1b describes the proposed DS-CSME network. The network accepts coil images computed from the ACS data as its inputs. The CSM estimation problem is naturally expressed as operating on coils set rather than a tensor of multichannel data: the number of coils is not fixed in advance. The coils have no inherent ordering. The proposed DS-CSME is designed to be equivariant to permutations by being built as a combination of deep elementwise functions and atomic reductions similar to the DeepSets architecture for permutation invariance [4]. The proposed DS-CSME uses the root-sum-of-squares (RSS) instead of the sum for reduction and injects it back into the coils by RSS normalization so that the output CSMs are normalized. The DS-CSME network is a stack of DeepSet blocks, each block being composed of a typical single-coil CNN applied to every coil followed by RSS normalization.
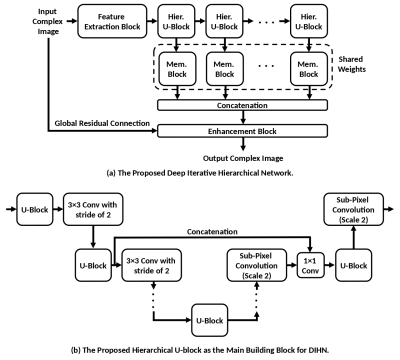
A new image-to-image translation network, referred to as deep iterative hierarchical network (DIHN), is introduced as the main building block of the proposed DS-CSME. As shown in Figure 2, DIHN extends the down-up network [5] with a novel hierarchical design that repeatedly decreases and increases the feature maps' resolution, allowing for a more efficient model as compared to conventional U-Nets.
Materials and Experimental Setting
Experiments were performed on 2D multi-coil slices acquired using various clinical 1.5T and 3T scanners with a fully sampled TSE sequence for all body, contrasts, orientations, SNR, etc. (9839 2D slices for training, 801 2D slices for testing). From this fully sampled data, retrospective downsampling was applied with an equispaced sampling mask with PAT 4, 75% phase resolution, and 16 fully sampled low-frequency reference lines. The proposed DS-CSME is compared against a similar reconstruction network using precomputed CSMs estimated using ESPIRiT [1]. The models were trained using a mixed loss of L1 and a multi-scale version of structural similarity (MS-SSIM). The CSM estimation network was first pre-trained to reproduce the precomputed CSM, then fine-tuned end-to-end and the reconstruction network's training.Results
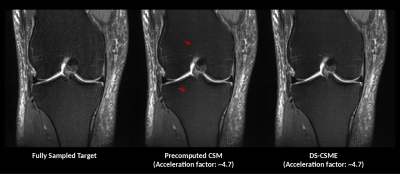
Compared to the fully sampled ground truth, predictions from the proposed end-to-end system with DC-CSME achieved a PSNR of 33.93 dB and SSIM of 0.840, like those obtained using precomputed CSMs. (PSNR of 33.95 dB and SSIM of 0.844). However, we observed more artifacts with precomputed CSMs, as shown in Figure 3. DS-CSME system required less time (~0.2s) to estimate CSMs than our ESPIRiT implementation (~1s).Conclusion
We introduced a deep sets approach for estimating CSMs that enables end-to-end DL networks for accelerated multi-coil MRI reconstruction. The proposed methods showed similar quantitative metrics to precomputed CSMs while offering improvements in performance speed and artifact reduction.Disclaimer
The concepts and information presented in this abstract/paper are based on research results that are not commercially available. Future availability cannot be guaranteed.Acknowledgements
No acknowledgement found.References
[1] Uecker M, Lai P, Murphy MJ, Virtue P, Elad M, Pauly JM, Vasanawala SS, Lustig M. ESPIRiT—an eigenvalue approach to autocalibrating parallel MRI: where SENSE meets GRAPPA. Magnetic resonance in medicine. 2014 Mar;71(3):990-1001.
[2] Uecker M, Hohage T, Block KT, Frahm J. Image reconstruction by regularized nonlinear inversion—joint estimation of coil sensitivities and image content. Magn. Reson. Med. 2008; 60:674–682.
[3] Holme, H.C.M., Rosenzweig, S., Ong, F. et al. ENLIVE: An Efficient Nonlinear Method for Calibrationless and Robust Parallel Imaging. Sci Rep 9, 3034 (2019).
[4] Zaheer M, Kottur S, Ravanbakhsh S, Poczos B, Salakhutdinov RR, Smola AJ. Deep sets. In Advances in neural information processing systems 2017 (pp. 3391-3401).
[5] Yu S, Park B, Jeong J. Deep iterative down-up CNN for image denoising. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops 2019 (pp. 0-0).
Figures