1162
MGRAPPA: Motion Corrected GRAPPA for MRI1Department of Mathematics, University of Maryland at College Park, College Park, MD, United States, 2Department of Diagnostic Radiology and Nuclear Medicine, University of Maryland School of Medicine, Baltimore, MD, United States, 3Center for Scientific Computation and Mathematical Modeling, University of Maryland at College Park, College Park, MD, United States
Synopsis
We introduce an approximation and resulting method called MGRAPPA to allow high speed MRI scans robust to subject motion using prospective motion correction and GRAPPA [1,2]. In experiments on both simulated data and in vivo data, we observe high accuracy and robustness to subject movement in L2 (Frobenius) norm error including a 41% improvement in the in vivo experiment.
Introduction
Subject motion is inevitable in MRI. Motion artifacts can be partially corrected using post-processing or prospective correction methods [3]. Here, we consider motion-induced image reconstruction errors for parallel imaging (PI). Calibration data are often acquired in the beginning of the entire imaging process. When motion occurs later, the pre-motion calibration data no longer match the spatial position and the corresponding coil sensitivity encoding of the subsampled data. The purpose of this study is to update reconstruction coefficients, with a focus on k-space based PI reconstruction using the Generalized Autocalibrating Partially Parallel Acquisition (GRAPPA) algorithm [1,2]. A similar concept has been extended to the image-based reconstructions using the Sensitivity Encoding (SENSE) method [4,5].With prospective motion correction (PMC), raw data are properly aligned in k-space, but subsequent GRAPPA reconstructions can show image artifacts since RF coils are stationary relative to the moving object. The problem can be solved by reacquiring GRAPPA kernels and re-scanning calibration data, but this is commonly impractical and requires additional scan time. Our new method, which we call MGRAPPA, recalculates GRAPPA kernels using motion tracking data to create a more stable and accurate reconstruction.
Theory
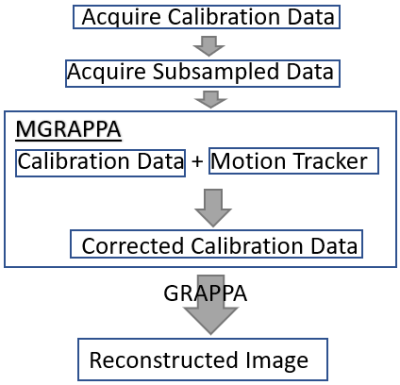
Figure 1 shows a flowchart of MGRAPPA in a typical PI scan. For each RF coil, a fully sampled calibration data set (centered in k-space) and an under-sampled data set are acquired. GRAPPA recovers missing k-space data by convolving the sub-sampled data with kernels calculated from calibration data. For MGRAPPA, the acquisitions remain unchanged, but the GRAPPA kernels are updated by reconstructing low-resolution coil sensitivity maps from the calibration data, transforming sensitivity maps inversely to subject motion, and recalculating calibration data and finally GRAPPA kernels.From each coil, we get $$$P_i^{0,0,0}f$$$ in the form of a matrix. First, we approximate the low frequencies of the signal, $$$f$$$, (ex: brain) by
$$L(f(x,y)) \approx \tilde f^l(x,y):=\sum_{i=1}^n | L(P_i^{0,0,0} f(x,y))|.$$
The approximation $$$\tilde f^l(x,y)$$$ is reasonable because the coil profiles, $$$P$$$, (superscripts are rotation, $$$\theta$$$, and translation, $$$x,y$$$, of coil profile and subscript is coil profile index) are approximately a partition of unity and lowpass, denoted $$$L$$$ ($$$L$$$ clears high frequency coefficients of the FFT), is linear. Set $$$\epsilon>0$$$ small and then approximate $$$P_i^{0,0,0}$$$ by
$$P_i^{0,0,0}(x,y) \approx \tilde P_i^{0,0,0}(x,y):=L(P_i^{0,0,0} f(x,y))/(\tilde f^l(x,y)+\epsilon \max_{x,y} | \tilde f^l(x,y)|).$$
The approximation $$$\tilde P_i^{0,0,0}(x,y)$$$ is reasonable because the profiles are very smooth. Then this implies that the high frequencies of $$$f$$$ in the numerator get removed by $$$L$$$ and then the low frequencies of $$$f$$$ in the numerator get removed by division with approximate low frequencies of $$$f$$$ denoted $$$\tilde f^l$$$ where $$$\epsilon$$$ is added to avoid division by 0 and increase robustness. Then
$$P_i^{\theta,x_0,y_0}(x,y)\approx\tilde P_i^{\theta,x_0,y_0}(x,y)=\tilde P_i^{0,0,0}(R_\theta(x,y)+(x_0,y_0)).$$
So now we have the matrices $$$\tilde f^l$$$ and $$$\tilde P_i^{\theta,x_0,y_0}$$$. Take the product and use the approximation $$$P_i^{\theta,x_0,y_0} f \approx \tilde P_i^{\theta,x_0,y_0} \tilde f^l $$$. The frequencies in the calibration region of this approximation is used to compute the new GRAPPA kernel and then reconstruct the subsampled image, see algorithm/pseudocode in Figure 2.
Simulation Method
Simulation k-space data was synthesized based on coil sensitivity maps calculated from a phantom experiment and clinical 3-D MP-RAGE coil-combined brain images previously acquired. The phantom experiment was conducted on a 3T Prisma scanner (Siemens,Erlangen,Germany) using a 20-channel head coil and a spherical water phantom (3-D MP-RAGE; FOV = 240×240$$$mm^2$$$, resolution = 0.9×0.9×0.9$$$mm^3$$$, 256×256×256, 256 slices, TR/TE/TI = 2600/2.29/1350ms, flip angle $$$8^{\circ}$$$). The human MP-RAGE brain images were acquired on a 3T TrioTim scanner (Siemens) (FOV 256×256$$$mm^2$$$, resolution 1×1×1$$$mm^3$$$, 256×256×160, 160 slices, TR/TE/TI = 2600/4.47/1000ms, flip angle $$$12^{\circ}$$$).In Vivo Experiment
Brain MRI was performed in a healthy volunteer (after obtaining consent) who was trained to rotate the head by $$$7^{\circ}$$$ to the left upon instruction in the middle of the scan. This experiment was conducted on a 3T Prisma with a 20-channel head coil and MP-RAGE sequence with FOV 240×240$$$mm^2$$$, resolution 1.9×1.9×1$$$mm^3$$$, 128×128×224, 224 slices, TR/TE/TI=2300/2.29/900ms, flip angle $$$8^{\circ}$$$.Simulation Results
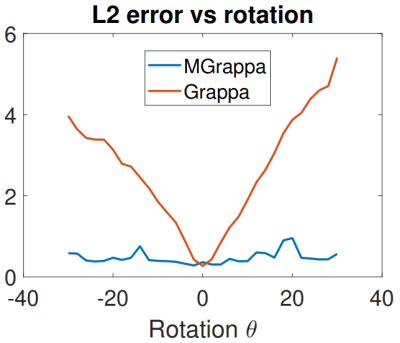
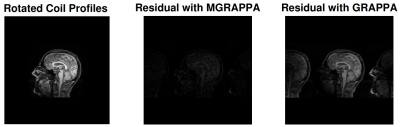
When rotation occurs with PMC, the apparent effect is inverse rotation of coil profiles by the same amount. In this experiment, we use a brain scan as the signal, and we use scans of a disk as profiles. Simulation parameters were: 20 coils, full resolution 256x256, calibration data 24*2 lines and $$$\epsilon=.05$$$. We simulate rotation of the profiles by calculation using various $$$\theta$$$ angles and then normalize the reconstructions in sup norm. We plot reconstruction errors for GRAPPA and MGRAPPA, see Figure 4. Error is L2 norm of the residual images; see Figure 3.In Vivo Results
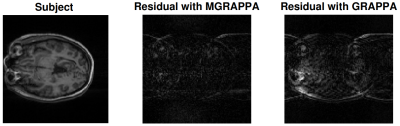
We collected full scans, 128x128, of a subject in 2 poses. We used the GRAPPA calibration data, 24*2 lines, from the first pose to reconstruct the scan from the second pose (subsample every other line and then reconstruct), both with and without MGRAPPA correction ($$$\epsilon=.05$$$). The L2-norm error decreases 41% with MGRAPPA, see Figure 5.Discussion
While the reconstruction error with GRAPPA increases markedly with rotations in an almost linear fashion, MGRAPPA uses approximations to mitigate this and results in a stable method with consistently low errors. In vivo, this appears as less ringing and blurring in the brain image We conclude that our method has great potential to reduce re-scanning and improve image quality in the presence of motion.Acknowledgements
This work was supported by NIH grant 1R01 DA021146 (BRP). Radu Balan was partially supported by NSF grant DMS-1816608.
References
[1] Mark A Griswold, Peter M Jakob, Robin M Heidemann, Mathias Nittka,Vladimir Jellus, Jianmin Wang, Berthold Kiefer, and Axel Haase. Generalized autocalibrating partially parallel acquisitions (grappa).Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine, 47(6):1202–1210, 2002.
[2] Ze Wang, Jiongjiong Wang, and John A Detre. Improved data reconstruction method for grappa. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine, 54(3):738–742, 2005.
[3] Julian Maclaren, Michael Herbst, Oliver Speck, and Maxim Zaitsev. Prospective motion correction in brain imaging: a review. Magnetic resonance in medicine, 69(3):621–636, 2013.
[4] Klaas P Pruessmann, Markus Weiger, Markus B Scheidegger, and Peter Boesiger. Sense: sensitivity encoding for fast mri. Magnetic Resonancein Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine, 42(5):952–962, 1999.
[5] Roland Bammer, Murat Aksoy, and Chunlei Liu. Augmented generalized sense reconstruction to correct for rigid body motion. Magnetic Resonance in Medicine: An Official Journal of the International Society for Magnetic Resonance in Medicine, 57(1):90–102, 2007.
Figures