1143
Registration-based whole breast segmentation enables highly reproducible quantitative MR-based breast density
Jia Ying1, Renee Cattell1, and Chuan Huang1,2,3
1Biomedical Engineering, Stony Brook University, Stony Brook, NY, United States, 2Radiology, Stony Brook Medicine, Stony Brook, NY, United States, 3Psychiatry, Stony Brook Medicine, Stony Brook, NY, United States
1Biomedical Engineering, Stony Brook University, Stony Brook, NY, United States, 2Radiology, Stony Brook Medicine, Stony Brook, NY, United States, 3Psychiatry, Stony Brook Medicine, Stony Brook, NY, United States
Synopsis
Clinical determination of mammographic breast density (MD) is subjective and can also suffer from intra- and inter-reader variability. MR-based quantitative assessment of breast density has a few unique advantages over mammography. However, accurate whole breast segmentation is crucial. Manual whole breast segmentation is burdensome and current automated methods also suffer from weaknesses. In this work, a new whole breast segmentation strategy based on image registration is proposed. The test-retest reliability of breast density derived using different segmentation methods was quantitatively assessed. The results demonstrated this new segmentation method yields more reproducible values compared to existing methods.
Purposes
Clinical determination of mammographic breast density (MD) according to the Breast Imaging Reporting and Data System (BI-RADS) is subjective and can also suffer from intra- and inter-reader variability [1, 2]. Volumetric MR-based technique is increasingly used in research due to some unique advantages including no ionizing radiation, no breast compression, high-resolution 3D capability and quantitative assessment [3]. Previous work has developed an MR-based breast density estimation method (MagDensity) [4] based on fat-water decomposition and breast segmentation, which is comparable to MD and is highly reproducible. However, accurate whole breast segmentation is essential for this task. Dynamic programming-based and deep learning-based breast segmentation approaches [4, 5] have difficulties in distinguishing pectoral muscles (lower boundary) that are jutting into the breast regions; thus, including the muscle regions in the breast segmentation, which impacts the accuracy of the MagDensity. Therefore, in order to improve the accuracy and reproducibility of MagDensity, a registration-based whole breast segmentation strategy is proposed.Methods
Data used in this work were obtained from healthy volunteers and patients with breast cancer. Axial mDixon-like images were acquired on 3T Siemens scanners in a multi-echo gradient echo fashion with 6 echoes. Complex images were saved for fat water separation. The breasts were divided in the middle, and the left breasts were flipped to the right. Thus, all breasts were processed as right-side breasts. Fifteen single-side breasts were chosen to form the dictionary of the template images. Manual segmentation was carefully performed on the template images according to Rosado-Toro J, et al [6].To remove artifacts and background noise, and correct intensity inhomogeneity, Canny edge detection, Otsu thresholding and MICO bias field correction method [7] were performed on both template images and new incoming images. The algorithm is summarized in Figure 1. First, normalized cross-correlation (NCC) was used to measure the similarity between the subject and each template image. The most similar 5 template images were chosen to perform global and non-rigid registrations individually using NiftyReg [8]. This was performed iteratively until the corresponding NCC value is greater than 0.9. The deformation map was also applied to the associated manual segmentation of the template image to generate registered mask. Only voxels contained in 4 or more registered template masks were included in the final segmentation of the breast.
Fifteen data sets with test-retest scans were used for evaluation. The test-retest scans were performed by repeating each scan by the same technologist on the same day. After the first scan, patients were taken out of the scanner and put back on the scanner immediately. Three subjects had 2 test-retest scan sessions (6 months apart), resulting in a total of 18 test-retest scan pairs. Only the unaffected breast (without cancer) was used in the analysis. The registration-based breast segmentation algorithm was applied to all scans (as subjects). Test-retest reproducibility of MagDensity for unaffected side of the breasts was assessed across four different segmentation methods: manual segmentation, dynamic programming-based segmentation, deep learning-based segmentation, and registration-based segmentation. The robustness was statistically evaluated using intraclass correlation coefficient (ICC) (two-way mixed effects model, single rater, absolute agreement) with 95% confidence interval.
Results
The test-retest reproducibility of MagDensity across four different segmentation methods are summarized in Table 1 and Fig. 2.Discussion
Among the four breast segmentation methods, the newly proposed registration-based method showed the highest ICC of 0.980, which indicated best test-retest reliability among these methods. Moreover, the registration-based method had shown the best correlation to the manual segmentation (the reference standard) with a Pearson correlation coefficient of 0.986 and yielded smallest variability during test-retest analysis compared with the other two automated ones (dynamic programming-based and deep learning-based).Conclusion
Our newly developed registration-based breast segmentation method yielded the best reproducibility in the task-based test-retest analysis among the 4 whole breast segmentation methods. It also had the best agreement with manual segmentation in terms of the estimated MagDensity.Acknowledgements
This work was in part supported by NIH grant R03CA223052.References
- Kerlikowske, K., et al. (1998). Variability and accuracy in mammographic interpretation using the American College of Radiology Breast Imaging Reporting and Data System. Journal of the National Cancer Institute, 90(23), 1801-1809.
- Ooms, E. A., et al. (2007). Mammography: interobserver variability in breast density assessment. The Breast, 16(6), 568-576.
- Wang, J., et al. (2013). Agreement of mammographic measures of volumetric breast density to MRI. PloS one, 8(12), e81653.
- Ding, J., et al. (2018). Reproducible automated breast density measure with no ionizing radiation using fat‐water decomposition MRI. Journal of Magnetic Resonance Imaging, 48(4), 971-981.
- Ding, J., Spuhler, K., & Huang, C. (2019). Deep learning-based whole breast segmentation for automated breast density measurements using fat-water decomposition MRI. 8th JSM of RCR & HKCR and 27th ASM of HKCR 2019.
- Rosado-Toro, J. A., et al. (2015). Automated breast segmentation of fat and water MR images using dynamic programming. Academic radiology, 22(2), 139-148.
- Li, C., Gore, J. C., & Davatzikos, C. (2014). Multiplicative intrinsic component optimization (MICO) for MRI bias field estimation and tissue segmentation. Magnetic resonance imaging, 32(7), 913-923.8.
- Modat, M., McClelland, J., & Ourselin, S. (2010). Lung registration using the NiftyReg package. Medical image analysis for the clinic-a grand Challenge, 2010, 33-42.
Figures
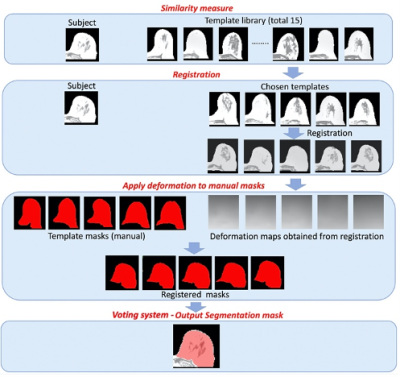
Figure 1. Pipeline for registration-based breast segmentation algorithm
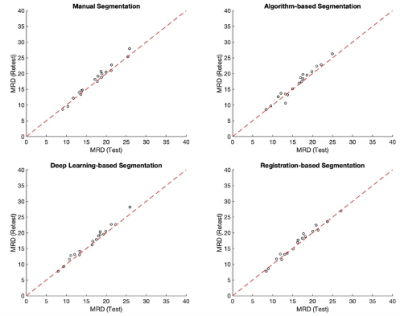
Figure 2. Comparison of test-retest reproducibility of MagDensity across four different segmentation methods.
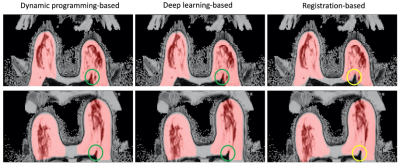
Figure 3. Registration-based breast segmentation method better identifies the lower boundary of the breast region. The resulting segmentations (red) are overlaid to the breast images with the problematic area circled in green.
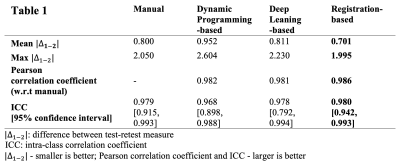
Table 1. Comparison of test-retest reproducibility of MagDensity across four different segmentation methods.