1117
Characterization of Breast Tumor using Machine Learning based upon Multi-parametric MRI Features.1Centre for Biomedical Engineering, Indian Institute of Technology Delhi, New Delhi, India, Delhi, India, 2Department of Radiology, Fortis Memorial Research Institute, Haryana, Gurgaon, India, Delhi, India, 3Department of Biomedical Engineering, All India Institute of Medical Science, New Delhi, India, Delhi, India
Synopsis
Multi-parametric MRI(mp-MRI) has shown promising outcomes with high sensitivity and accuracy in the characterization of breast tumor. Quantitative analysis of mp-MRI and texture features with machine learning approach have also shown potential in improving accuracy of breast tumor classification. The objective of this study was to differentiate low-grade vs. high-grade breast tumor using machine learning with optimized feature vector obtained from mp-MRI data. The study included mp-MRI data of 35 patients with breast cancer. The combination of support-vector-machine(SVM) with Wrapper method using Adaptive-Boosting(AdaBoost) technique resulted in high sensitivity(0.94±0.07), specificity(0.80±0.05), and accuracy(0.90±5.48) in classification of low-grade vs. high-grade tumors.
Introduction:
MRI can be used to obtain multi-parametric(mp) information such as structural, hemodynamic, physiological, etc1,2. Multiple studies have been reported focusing on the use of mp-MRI data for the characterization of breast tumor with varying sensitivity and specificity3,4,5. Radjenovic et al. have reported pharmacokinetic parameters from DCE-MRI for grading of invasive breast tumors. The volume transfer coefficient(Ktrans) and rate constant(Kep) were significantly higher in grade-III tumors than in grade-II tumors. None of the measured parameters varied significantly between grade-I and grade-II tumor3. Jiang et al. reported sensitivity of 57% and specificity of 78% with morphology features4. The quantitative analysis of mp-MRI data has shown potential in improving accuracy of breast tumor classification2. In general, a large set of quantitative and texture features can be generated depending upon the type of methodology used. A suitable combination of selected quantitative and texture features can further improve accuracy of tumor classification. Machine learning(ML) classifiers based upon features derived from MRI data has shown potential in the classification of tumors6,7. There is a need for further research studies on selection of appropriate combination of features as well to evaluate performance of different ML classifiers for accurate classification of breast tumor. In the current study, it was hypothesized that quantitative features(tracer kinetic, hemodynamic and diffusion) in combination with some texture features can improve accuracy of tumor classification using an optimized ML classifier. The objectives of the current study were to develop and optimize an machine learning framework for characterization of breast tumor(low-grade vs. high-grade tumor) using an optimized feature vector computed from mp-MRI data.Methods:
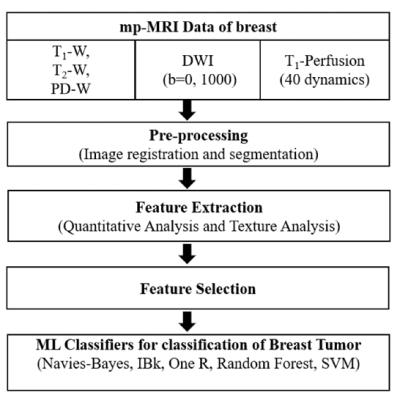
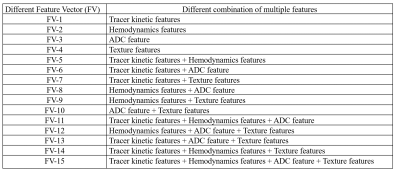
All the MRI experiments were performed at 3T-whole body Ingenia MRI system(Philips Healthcare,The Netherlands) using a 7-channel biopsy compatible breast coil. This study included breast mp-MRI data of 35-female patients with histopathology results. Protocol included conventional-MRI(T1-W,T2-W, and PD-W), Diffusion-weighted-imaging(DWI) with different b-values(0,1000s/mm2) and T1-perfusion MRI data with 40-dynamics, 5.4-seconds temporal-resolution).Data processing: The mp-MRI data was processed for extraction of features followed by selection of features and evaluation of multiple ML classifiers for classification of breast tumors(Figure-1). Tumor tissue was used as a region of interest(ROI). Quantitative features and texture features were extracted from ROI:1) 4-Tracer kinetics features(Ktrans , Kep, Vp, Ve) and 3-hemodynamic features(BBV, BBVcorr, and BBF), computed from T1-perfusion MRI data5, 2) apparent-diffusion-coefficient(ADC) parameter, computed using DWI data8, and 3) 40-texture features9,10 were computed from each 3 T1-perfusion images(pre-, peak- and delayed-contrast) using Radiomics tool. A total 128 features(8-quantitative features and 120-texture features) per patient were used in this study. Finally, a 15-feature vectors(FVs) consists of different combinations of quantitative and texture features were used in this study as shown in Table-1. Wrapper-based-feature-selection(WBFS) method11with Adaptive-Boosting technique was used for feature selection. 5-ML classifiers were evaluated for tumor classification using 10-fold cross-validation(CV) with 10-repetitions. The diagnostic performance for differentiating between low-grade vs. high-grade tumors using different classifiers without and with the feature selection was analyzed using accuracy, sensitivity, specificity, precision, F1-score, mean-absolute-error(MAE) and root-mean-squared-error(RMSE) with respect to histology result.
Results:
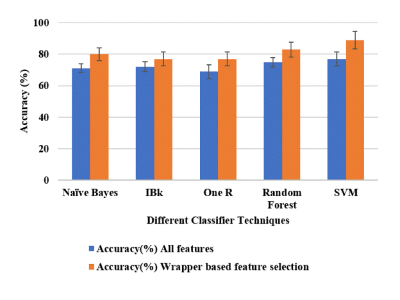
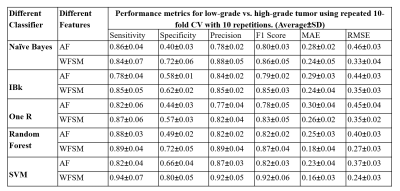
The performance of 5 different classifiers and 15 different FVs, without and with feature selection, were evaluated for classification of low-grade vs. high-grade tumor. Among all FVs, the FV-15 feature vector without any feature selection, with SVM classifier provided highest average accuracy(77%±4.31) and AUC(0.76±0.06) for the classification of low-grade vs. high-grade tumors with 10-fold CV over 10-repetitions. WBFS techniques provided reduced number of features in this feature vector (FV-15). WBFS method selected 5 features(BBV, BBVcorr, Correlation at peak-contrast, Gray-Level Variance(GLV) from GLRLM at delayed contrast and High Gray Level Zone Emphasis(HGZE) from GLSZM at delayed contrast to classify the low-grade vs. high-grade tumor. SVM with optimum features selected using Wrapper method with Adaptive-Boosting technique provided a highest average accuracy(90%±5.48) and AUC(0.93±0.10) as shown in Figure-2. Table-2 shows that the SVM classifier with the WBFS method provided the highest sensitivity(0.94±0.07), specificity (0.80±0.05), precision(0.92±0.05), and F1 score(0.92±0.06) in characterizing the tumor between low-grade vs. high-grade tumors among all other methods using 10-fold CV method over 10-repetitions. It also provided the lowest MAE(0.16±0.03) and RMSE(0.24±0.03).Discussion and Conclusion:
The choice of features plays a crucial part in the accurate classification of tumors. In the current study, hemodynamic features were combined with other features and used for ML classifier development. Hemodynamic features are biomarkers for angiogenesis, which has been reported to be correlated with tumor growth and tumor type5. Inclusion of hemodynamic features in mp-MRI feature vector improved accuracy of tumor classification substantially. The AdaBoost method reduced the problem of imbalance data and mitigated the problem of overfitting. Using a combination of SVM and optimal mp-MRI features set with wrapper method provided the highest sensitivity, specificity and accuracy as compared to reported studies3,4,5. These are preliminary results with a small number of patients. It is necessary to validate the classifier on a larger data size in future studies. In conclusion, hemodynamic features were included with tracer kinetic, ADC and texture features to create a feature vector in the current study, which improved the accuracy in the classification of low vs. high grade tumor. Our finding provided that the SVM outperformed other ML methods in the binary classification of breast tumor.Acknowledgements
The authors thank Dr. Sunita Ahlawat for providing histopathology results; Rupsa Bhattacharjee for technical assistance. The Authors acknowledge an internal grant from the Indian Institute of Technology Delhi.References
1. Rahbar H, Partridge SC. Multiparametric Breast MRI of Breast Cancer. Magn Reson Imaging Clin N Am. 2016;24(1):223-238.
2. Pinker K, Helbich TH, Morris EA. The potential of multiparametric MRI of the breast. Br Inst Radiol. 2017;90(20160715):1-17.
3. Radjennovic A, Dall BJ, Ridgway JP, Smith MA. Measurement of pharmacokinetic parameters in histologically graded invasive breast tumours using dynamic contrast-enhanced MRI. Br J Radiol. 2008;81(962):120-128.
4. Jiang X, Xie F, Liu L, Peng Y, Cai H, Li LI. Discrimination of malignant and benign breast masses using automatic segmentation and features extracted from dynamic contrast-enhanced and diffusion-weighted mri. Oncol Lett. 2018;16(2):1521-1528.
5. Thakran S, Chatterjee S, Singhal M, Gupta RK. Automatic outer and inner breast tissue segmentation using multi-parametric MRI images of breast tumor patients. Published online 2018:1-21.
6. Cai H, Peng Y, Ou C, Chen M, Li L. Diagnosis of breast masses from dynamic contrast-enhanced and diffusion-weighted MR: A machine learning approach. PLoS One. 2014;9(1).
7. Liu B, Li X, Li J, et al. Comparison of Machine Learning Classifiers for Breast Cancer Diagnosis Based on Feature Selection. Proc - 2018 IEEE Int Conf Syst Man, Cybern SMC 2018. Published online 2019:4399-4404.
8. Woodhams R, Matsunaga K, Iwabuchi K, Hata H, Kuranami M, Hayakawa K. Diffusion-Weighted Imaging of Malignant Breast Tumors The Usefulness of Apparent Diffusion Coefficient (ADC) Value and ADC Map for the Detection of Malignant Breast Tumors and Evaluation of Cancer Extension. J Comput Assist Tomogr. 2005;29(5):644-649.
9. Agner SC, Soman S, Libfeld E, et al. Textural kinetics: A novel dynamic contrast-enhanced (DCE)-MRI feature for breast lesion classification. J Digit Imaging. 2011;24(3):446-463.
10. Haralick R, Shanmugam K, and Dinstein I, Textural Features for Image Classication, IEEE Transactions on Systems, Man, and Cybernetics, vol. SMC-3, no. 6, pp. 610-621, Nov. 1973.
11. Darzi M, AsgharLiaei A, Hosseini M, HabibollahAsghari. Feature Selection for Breast Cancer Diagnosis: A Case-Based Wrapper Approach. World Acad Sci Eng Technol. 2011;77(5):1142-1145.
Figures