1114
Predicting Underestimation of Invasive Cancer in Patients with Core-needle Biopsy-diagnosed Ductal Carcinoma in Situ using Deep Learning1Department of Radiology, Chonnam National University, Gwangju, Korea, Republic of, 2College of Medicine, Chonnam National University, Gwangju, Korea, Republic of, 3Department of Radiology, Chonnam National University Hwasun Hospital, Hwasun, Korea, Republic of, 4Department of Artificial Intelligence Convergence, Chonnam National University, Gwangju, Korea, Republic of, 5Department of Radiology, Chonnam National University Hospital, Gwangju, Korea, Republic of
Synopsis
This study aims to explore the effectiveness of deep learning algorithms for distinguishing pure (noninvasive) ductal carcinoma in situ (DCIS) from invasive disease for patients showing DCIS in core-needle biopsy using MRI. Preoperative axial dynamic contrast-enhanced MRI data from 352 patients were used to train, validate and test the two-step convolutional neural network (CNN) utilizing a recurrent model. Our model produced an accuracy of 69.4% and AUC of 0.721. The comparison between the proposed model and a 2D or 3D model suggests that the sequential information may provide an important support for occult invasive cancer in patients diagnosed with DCIS.
PURPOSE
The prediction of occult invasive disease in DCIS before surgery is of great importance for providing more personalized and proper treatment strategy. We demonstrated the potential of recurrent deep learning model for identifying invasiveness in DCIS diagnosed by core needle biopsy.METHOD AND MATERIALS
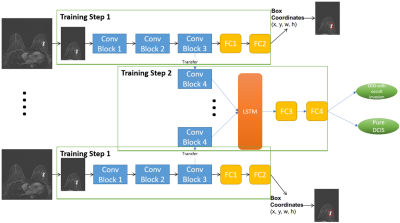
In this retrospective study, a total of 352 patients diagnosed with DCIS by core needle biopsy from 2011 to 2017 were included. Preoperative axial T1-weighted dynamic contrast-enhanced MRI was acquired using a clinical 3T scanner, and the subtraction images between pre- and post-contrast imaging sequences were utilized. The data were selected based on the assumption that only one side of the breast contains DCIS. The ground-truth labels pure DCIS (n=202) and DCIS with occult invasion (n=150) were identified based on the final pathology report and ROIs of DCIS drawn manually by an experienced radiologist. The datasets were divided into training (n=220), validation (n=60) and testing (n=72). A standard normalization was applied for the entire slices of each data sample. One quarter of the image that contained DCIS was automatically cropped and used for training with deep learning algorithms. The training data were constructed by using the first three subtraction sequences and augmented by rotation, horizontal and vertical flip.A CNN-based algorithm was developed to train the classifier model for the binary classification of pure DCIS and DCIS with occult invasion. The input of our models was a 20-slices sequence of 3-channels DICOM images. Three channels corresponded to the 1st, 2nd and 3rd subtraction image, respectively. A 2-step algorithm was implemented to account for the difference in the size of tumors among patients. First, we designed a CNN network for the task of localizing the bounding box containing breast tumor for a single imaging slice. A regression model was attached to the end of this CNN to produce the coordinates of tumor ROIs. Second, we fine-tuned the CNN network in the first step by adding one more convolution block, a LSTM layer, and replacing the regression model with Softmax classifier. The LSTM layer was utilized for capturing the sequential information through multiple MR imaging slices. The detailed diagram of the proposed algorithm is described in Figure 1. The CNN network in the first step was trained to perform the localization task and at the same time, served as a pre-training model for the classification network in the second step.
In order to explore the effectiveness of the proposed method of using the recurrent model for multi-slice MRI data, we applied a 2D-CNN and 3D-CNN model by considering each image slice as a single data sample and by using multi-slice images as a 3D input, respectively. In the second step, instead of using LSTM layer, we adjusted one more convolution block. The evaluation of 2D model is implemented by considering the slice with highest score as the final outcome of one patient data.
RESULTS AND DISCUSSIONS
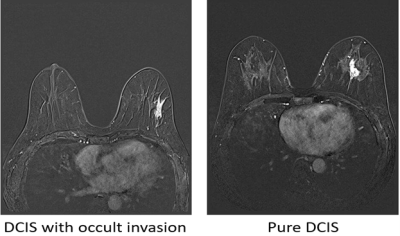
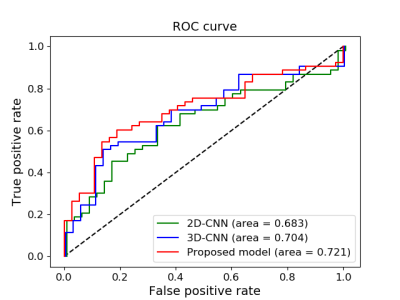
Figure 2 exhibited examples of subtraction images from pure DCIS and DCIS with occult invasion. Table 1 shows the comparison of results between the proposed 2-step model, 2D-CNN, and 3D-CNN model for the classification of pure DCIS versus DCIS with occult invasion. The F1-score, accuracy, and AUC and of the proposed model for the validation data were 0.71, 68.3% and 0.708, respectively, which were similar to those of 3D-CNN, but higher than those of 2D CNN model. For the testing data, the proposed model achieved the level of performance (F1-score, 0.71; accuracy, 69.4%; AUC, 0.721) that were higher than those of other models. The performance of 2D-CNN and 3D-CNN models were similar in the testing dataset. Figure 3 shows ROC curves for the three models.A previous study on distinguishing pure DCIS from DCIS with occult invasion suggested that the hand-crafted MRI features [1] and 2D deep learning model [2] can achieve promising results. In this paper, we developed a 2-step algorithm utilizing a recurrent CNN model and demonstrated that the proposed algorithm can provide a method to predict invasiveness in the core needle biopsy-proven DCIS with the results comparable to the previous reports [1,2].
Conclusion.
The prediction of occult invasive disease in DCIS before surgery is of great importance for providing more personalized and proper treatment strategy. We demonstrated the potential of our model for identifying Invasiveness in DCIS diagnosed by core needle biopsy using subtracted T1 MRI images.Acknowledgements
This study was supported by the Ministry of Education, Republic of Korea (2019R1I1A3A01059201) and the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (HR20C0021).References
1. Harowicz MR, Saha A, Grimm LJ, et al., Can algorithmically assessed MRI features predict which patients with a preoperative diagnosis of ductal carcinoma in situ are upstaged to invasive breast cancer? J. Magn. Reson. Imaging (2017) 1–9.
2. Zhu Z, Harowicz MR, Zhang J, et al., Deep learning analysis of breast MRIs for prediction of occult invasive disease in ductal carcinoma in situ. Computers in Biology and Medicine 115 (2019).