1094
The Open Source Initiative for Perfusion Imaging (OSIPI): DCE-MRI Challenge
Anahita Fathi Kazerooni1, Laura C. Bell2, Floris Van den Abeele3, Ruben Verhack3, Xinze Zhou4, Salman Rezaei5, Zaki Ahmed6, Rianne Van Der Heijden7, Seyed Ali Nabavizadeh4, Leland S. Hu8, Hamidreza Saligheh Rad5, and Steven Sourbron9
1Department of Radiology, UPenn, Philadelphia, PA, United States, 2Division of Neuroimaging Research, Barrow Neurological Institute, Phoenix, PA, United States, 3Hyperfusion.ai, Gent, Belgium, 4University of Pennsylvania, Philadelphia, PA, United States, 5Quantitative MR Imaging and Spectroscopy Group, Research Center for Molecular and Cellular Imaging, Tehran University of Medical Sciences, Tehran, Iran (Islamic Republic of), 6Mayo Clinic, Rochester, MN, United States, 7Department of Radiology & Nuclear Medicine, Erasmus MC University Medical Center, Rotterdam, Netherlands, 8Neuroradiology Division, Department of Radiology, Mayo Clinic, Phoenix, AZ, United States, 9University of Sheffield, Sheffield, United Kingdom
1Department of Radiology, UPenn, Philadelphia, PA, United States, 2Division of Neuroimaging Research, Barrow Neurological Institute, Phoenix, PA, United States, 3Hyperfusion.ai, Gent, Belgium, 4University of Pennsylvania, Philadelphia, PA, United States, 5Quantitative MR Imaging and Spectroscopy Group, Research Center for Molecular and Cellular Imaging, Tehran University of Medical Sciences, Tehran, Iran (Islamic Republic of), 6Mayo Clinic, Rochester, MN, United States, 7Department of Radiology & Nuclear Medicine, Erasmus MC University Medical Center, Rotterdam, Netherlands, 8Neuroradiology Division, Department of Radiology, Mayo Clinic, Phoenix, AZ, United States, 9University of Sheffield, Sheffield, United Kingdom
Synopsis
Dynamic contrast enhanced (DCE-) MRI is widely acquired as a part of neuroimaging protocol for evaluation of glioblastoma tumors before the start of therapy or monitoring and assessment of treatment response in longitudinal scans. Nonetheless, lack of a standardized quantification method has limited its application in clinical settings, multi-institutional studies and clinical trials. These challenges have motivated efforts to validate DCE-MRI using benchmark biomedical image analysis methods. The Open-Source Initiative for Perfusion Imaging (OSIPI) has designed the OSIPI-DCE challenge to evaluate and compare DCE tools in terms of accuracy, repeatability, and reproducibility of Ktrans estimation in the brain.
Purpose
Research findings cannot be reproduced when researchers evaluate new methods on personal datasets and local code that is not shared with the public [1]. With a growing attention to reproducible research at ISMRM [2], community challenges with public datasets can play an important role in validating and benchmarking image quantification methods [3]. While dynamic contrast enhanced (DCE-) MRI has been widely used to evaluate a diversity of brain pathologies, there is a lack of standardized analysis methods, limiting its application in clinical practice, multi-institutional studies, or clinical trials. The Open-Source Initiative for Perfusion Imaging (OSIPI), an initiative of ISMRM perfusion study group and sponsored by ISMRM, was founded to promote reproducible research and open science in perfusion imaging and facilitate translation of software tools into clinical practice. In accordance with the aims of OSIPI, we have designed the DCE (OSIPI-DCE) challenge for evaluating DCE-MRI methods that estimate the volume transfer constant (Ktrans) in brain tumors.Challenge Overview
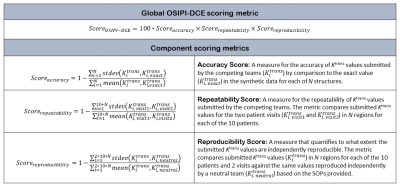
We will evaluate and compare the analysis methods submitted by the competing teams for quantification of Ktrans from DCE-MRI in terms of accuracy, repeatability, and reproducibility. The accuracy of Ktrans estimations by the participating teams will be scored based on synthetic data specifically designed for this challenge; repeatability will be rated based on test-retest DCE-MRI scans of 10 patients with glioblastoma brain tumor; reproducibility will be assessed based on an independent analysis by a neutral team. The submissions will be ranked according to a global score reflecting that an ideal method should be accurate AND repeatable AND reproducible (Table 1). The challenge will be advertised in http://challenge.ismrm.org, results will be announced in the Perfusion Study Group Workshop in February 2022, and the top three teams will present their methods at the ISMRM 2022 annual meeting. Teams interested in this challenge will be asked to register for free on http://challenge.ismrm.org. Upon registration, the teams will receive submission guidelines and a link to the data, which is uploaded in a repository. The challenge will run over a course of 9 months, but each competing group will be granted 3 months from the time of their registration to submit their reports and results (Figure 1). At the end of the challenge, all the submissions will be processed and evaluated by the organizing team.Data
The following data will be provided for the competing teams:- Synthetic: This synthetic data is designed based on a two-compartment exchange model (2CXM) and the signal model of a spoiled gradient-echo sequence in steady-state. It contains multiple structures with specifications of normal gray matter, normal white matter, tumor, necrosis, and arterial input function (AIF) taking a range of values for PS, Ve, Vp, and F. Temporal sampling time is 6 s, the total scan time is 6 min and Rician noise has been added. T1 values for AIF and tissue are 1400ms and 1000ms, respectively.
- Clinical: A set of test-retest DCE-MR images from 10 patients with newly diagnosed glioblastoma scanned on 3T scanners at two scan dates typically 2-5 days apart [4,5] will be provided (details of imaging parameters can be found in Table 2).
Submissions and evaluation
Competing teams will be required to submit (1) matrices of voxel-wise Ktrans maps for the synthetic and clinical data (2 visits per patient) in NIfTI format, and (2) Standard Operating Procedures (SOPs) that will allow a neutral team to reproduce the results. The SOPs should explain how to access and install the software used. There is no requirement to release the source code, but for commercial software or in-house software that is not freely available, a temporary license should be granted to the neutral team so the results can be reproduced. The submissions will be ranked according to the OSIPI-DCE score defined in Table 1.Discussion
The OSIPI-DCE challenge will provide a ranking of available software tools that will help future users select an appropriate method for their analysis. After the challenge is concluded, the data and procedures will be made freely available to serve as a benchmark for any future developments in Ktrans mapping in the brain. The expectation is that providing robust benchmarks will ultimately promote a more standardized approach to the analysis of DCE and provide an evidence base for a future consensus [6]. As such, this challenge can support the uptake of Ktrans as a biomarker in multi-institutional clinical trials, drug development and ultimately clinical practice.Acknowledgements
The authors would like to acknowledge Dr. Keyvan Farahani, from National Cancer Institute, USA, and Dr.Natenael Semmineh, from Barrow Neurological Institute, USA, for their help in providing information for the design of this challengeReferences
- Maier-Hein, L. et al. Why rankings of biomedical image analysis competitions should be interpreted with care. Nat. Commun. 9, 5217 (2018).
- Stikov, N., Trzasko, J. D. & Bernstein, M. A. Reproducibility and the future of MRI research. Magn. Reson. Med. 82, 1981–1983 (2019).
- Maier-Hein, L. et al. BIAS: Transparent reporting of biomedical image analysis challenges. Med. Image Anal. 66, 101796 (2020).
- Prah, M. A. et al. Repeatability of Standardized and Normalized Relative CBV in Patients with Newly Diagnosed Glioblastoma. Am. J. Neuroradiol. 36, 1654 LP – 1661 (2015).
- Clark, K. et al. The Cancer Imaging Archive (TCIA): Maintaining and Operating a Public Information Repository. J. Digit. Imaging 26, 1045–1057 (2013).
- Boxerman, J. L. et al. Consensus Recommendations for a Dynamic Susceptibility Contrast MRI Protocol for Use in High-Grade Gliomas. Neuro. Oncol. (2020) doi:10.1093/neuonc/noaa141.