1093
The Open Source Initiative for Perfusion Imaging (OSIPI): Contrast-based perfusion lexicon and reporting recommendations1Medical Image Computing, German Cancer Research Center, Heidelberg, Germany, 2Division of Neuroscience and Experimental Psychology, University of Manchester, Manchester, United Kingdom, 3Department of Infection, Immunity and Cardiovascular Disease, University of Sheffield, Sheffield, United Kingdom, 4Department of Radiation Oncology, Netherlands Cancer Institute, Amsterdam, Netherlands, 5Barrow Neurological Institute, Phoenix, AZ, United States, 6Department of Radiology, Erasmus MC, Rotterdam, Netherlands, 7Brigham and Women’s Hospital, Boston, MA, United States, 8Department of Radiation Physics, University of Gothenburg, Gothenburg, Sweden, 9Steinbuch Centre for Computing, Karlsruhe Institute for Technology, Karlsruhe, Germany, 10PixelMed Publishing, LLC, Bangor, PA, United States, 11University of Vienna, Vienna, Austria, 12Mayo Clinic, Rochester, MN, United States, 13University of Leeds, Leeds, United Kingdom
Synopsis
As part of the Open Source Initiative for Perfusion Imaging (OSIPI), the aim of this work is to develop guidelines for reporting of contrast-based perfusion analysis in order to improve reproducibility, reusability and interoperability of perfusion analysis. For this, a perfusion analysis lexicon is developed, which provides standardized nomenclature for perfusion parameters and analysis processes, as well as a framework for reporting of analysis pipelines. The lexicon is intended to be a dynamically growing inventory updated by the perfusion community. For that, a variety of public feedback and review cycles are planned during the development process.
Introduction
Dynamic contrast-enhanced (DCE) and dynamic susceptibility contrast (DSC) magnetic resonance imaging (MRI) are used to measure quantitative tissue perfusion properties. Perfusion parameters are extracted from the acquired images via a sequence of processes, which define an image analysis pipeline. Many software platforms exist (commercial, in-house, open-source) to analyze perfusion data; however, comparison of results between software platforms is difficult due to reporting differences and the inability to document the processes applied during analysis. Also there is no consistency in how analysis results are shared and how details are reported in publications. A key element in ensuring perfusion reproducibility is the standardization of nomenclature (i.e., naming parameters) and reporting of processing pipelines– both of which are currently lacking.As part of the Open Source Initiative for Perfusion Imaging (OSIPI)1, the main aim of this work is to develop guidelines for reporting to improve reproducibility of perfusion analysis. For this, a perfusion analysis lexicon was developed which provides standardized nomenclature for perfusion parameters and analysis processes. Additionally, a framework for reporting DCE and DSC analysis pipelines based on the lexicon is provided. In this abstract, we present the current status of the lexicon, the timeline for completion, and potential future applications.
Methods
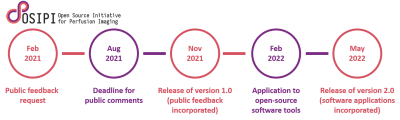
The perfusion lexicon was developed as part of OSIPI by task force (TF) 4.2 2, consisting of experts in the fields of DCE and DSC MRI as well as Digital Imaging and Communications in Medicine (DICOM) standardization experts.The main milestones and deliverables of the first 2-year cycle of the lexicon development are depicted in Figure 1. After being drafted by TF 4.2 and undergoing an internal OSIPI review, the lexicon will be available for public consultation (request e.g. via the ISMRM perfusion study group mailing list) in February 2021. Public comments will be closed in August 2021, and after incorporation of the feedback, version 1.0 of the lexicon will be released in November 2021.
It is planned in the second phase to extend the lexicon for compatibility with the processing pipelines implemented in a list of open-source perfusion analysis tools collected by OSIPI TF 1.2 3. Version 2.0 of the lexicon is planned to be released in May 2022.
Results
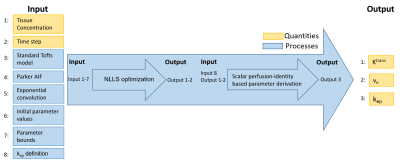
Lexicon structure: The lexicon is composed of different sections, each containing a list of items that are arranged in thematic groups. Each item is uniquely identified by a code and also assigned a human-readable name. An overview is provided in Figure 2.Quantities/processes: Two types of items are defined: quantities represent variables that can act as input/output to an analysis pipeline. Processes act as operators that generate output quantities based on given input quantities. They can also act as inputs to other processes, for instance forward models can be used as inputs to optimization procedures.
Primary/derived processes: Processes can be combined or composed to produce new processes (derived processes) to maximise opportunities for re-use and simplify the encoding of complex pipelines. These are stored in the section Use Cases. All other sections represent primary processes, i.e. processes that are not defined in terms of other processes.
Pseudocode: The lexicon provides a structured approach to writing pseudocode that can represent any DCE/DSC analysis pipeline. The pseudocode is semantic in the sense that it defines what variables and functions mean (i.e. physical interpretation) but not how they are represented (i.e. which data type). Table 1 and Figure 3 show an example of a derived process encoded using the lexicon.
The lexicon is a public document which can be found here: http://bit.ly/perfusion-reporting.
Discussion and Outlook
The lexicon is intended to be a dynamically growing inventory updated by the perfusion community and has been designed such that it can be easily extended. The scope of version 1.0 is defined by what was considered of primary importance by the TF members, an OSIPI internal review, and based on feedback from the first round of public consultations. In version 2.0, the scope will be extended to match the workflows implemented by open-source software tools. The long-term plan is to establish regular update phases leading to different lexicon versions.The lexicon recommends nomenclature and provides a reporting framework, but it does not give recommendations on how to perform the analysis itself, or what format should be used for representing analysis results. It is the aim, however, to align the lexicon with initiatives providing such recommendations. For example, the lexicon is able to describe the recommended analysis pipelines provided by the QIBA initiative 4,5.
A potential application of the lexicon is to amend the DICOM standard for the purposes of communicating parametric maps of contrast agent based perfusion MRI 6. This will facilitate data sharing of derived parametric maps by providing a structured communication of quantities, source data and metadata. An OSIPI task force (TF 4.3)7 is planned for this after version 1.0 has been released. The lexicon is also linked to OSIPI TF 2.3 which is using the structure and nomenclature of the lexicon to structure a code repository 8.
In summary, a lexicon is provided which can be used to standardize the nomenclature and reporting of perfusion analysis in order to improve the reproducibility, reusability and interoperability.
Acknowledgements
No acknowledgement found.References
1. https://www.osipi.org
2. https://www.osipi.org/task-force-4-2/
3. https://www.osipi.org/task-force-1-2/
4. QIBA MR Biomarker Committee. MR DCE-MRI Quantification (DCEMRI-Q), Quantitative Imaging Biomarkers Alliance. Profile Stage: Public comment. Dec 10, 2020. Available from: https://qibawiki.rsna.org/index.php/Profiles, accessed on: Dec 14, 2020.
5. QIBA MR Biomarker Committee. MR Dynamic Susceptibility Contrast (DSC), Quantitative Imaging Biomarkers Alliance. Profile Stage: Consensus. Oct 22, 2020. Available from: https://qibawiki.rsna.org/index.php/Profiles, accessed on: Dec 14, 2020.
6. Fedorov, A., Beichel, R., Kalpathy-Cramer, J., Clunie, D., Onken, M., Riesmeier, J., Herz, C., Bauer, C., Beers, A., Fillion-Robin, J.-C., Lasso, A., Pinter, C., Pieper, S., Nolden, M., Maier-Hein, K., Herrmann, M. D., Saltz, J., Prior, F., Fennessy, F., Buatti, J. & Kikinis, R. Quantitative Imaging Informatics for Cancer Research. JCO Clin Cancer Inform 4, 444–453 (2020).
7. https://www.osipi.org/task-force-4-3/
8. https://www.osipi.org/task-force-2-3/
Figures