1045
Lateralization of Temporal Lobe Epilepsy Using Multimodal MRI, Decision Tree, and Random Forest Methods1Shahed University, Tehran, Iran (Islamic Republic of), 2Tehran University of Medical Sciences, Tehran, Iran (Islamic Republic of), 3Brain and Spinal Cord Injury Research Centre, Neuroscience Institute, Tehran University of Medical Sciences, Tehran, Iran (Islamic Republic of), 4Biomedical Engineering Department, Shahed University, Tehran, Iran (Islamic Republic of), 5dr.mehvari@hotmail.com, Isfahan, Iran (Islamic Republic of)
Synopsis
In this study, a decision making method was developed for determination epileptogenicity in mesial temporal lobe epilepsy (mTLE) patients using different neuroimaging markers including hippocampal volume, and FLAIR (Fluid Attenuated Inversion Recovery) intensity and MD (Mean Diffusivity) value in hippocampus, FA (Fractional anisotropy) in posteroinferior cingulum, and FA in crus of fornix from MRI images of T1, FLAIR, and DTI (diffusion tensor imaging). The aim of this study is to creating an automated classification algorithm using decision tree and random forest methods. Result of applied method detected essential rules for prediction of laterality in individual mTLE patients.
Introduction
Mesial temporal lobe epilepsy (mTLE) is the most common type of pharmacoresistant focal epilepsy that patient candidate for surgical treatment 1,2 .The success of surgery depends on the accurate localization of seizure onset. In this study using multistructural MR images of T1, FLAIR (Fluid Attenuated Inversion Recovery), and DTI (Diffusion Tensor Imaging), we proposed a classification algorithm using decision tree method, which was applied for finding essential rules for prediction of laterality in individual mTLE patients. We also used random forest techniques for understanding the importance of extracted imaging attributes.Methods
Thirty-five unilateral mTLE patients who had candidates for surgical resection were studied prospectively. Hemispheric variation uncertainty (HVU) of hippocampal T1 volumetry and FLAIR intensity was estimated from structural MRI using a 64-channel phased-array head coil on a 3-Tesla scanner (Siemens Prisma, Erlangen, Germany) at Iranian National Brain Mapping Laboratory (NMBL). Anatomic images were acquired for clinical diagnosis using a standardized protocol including transverse T1 weighted images using MPRAGEIR protocol with the following imaging parameter: TR = 1840 ms, TI=900 ms, TE = 2.43ms, flip angle = 8°, matrix = 224 224, in-plane resolusion=1.0 1.0 ,slice thickness = 1.0 mm, pixel bandwidth = 250 Hz/pixel. HVU levels of mean diffusivity (MD) in the hippocampus and fractional anisotropy (FA) in posteroinferior cingulum and crus of fornix were also estimated from DTI. Multishell DTI images (b-values of 1000 and 2000 s/mm2)in 64 diffusion gradient directions on each shell along with a set of null images (b-value of 0 s/mm2) were acquired using echo planar imaging (EPI) onsame machine in anterior to posterior phase direction with the following imaging parameters: TR = 9600 ms, TE = 92 ms, flip angle = 90 , matrix = 110 110, in-plane resolusion = 2.0 .0 , slice thickness = 2.0 mm, pixel bandwidth = 1420 Hz/pixel. Each estimated HVU value was used as a ground to compare the interhemispheric change in the corresponding imaging attribute with and to establish a label of laterality ‘beyond the uncertainty’ for individual mTLE cases. Decision tree method was applied on laterality labels to provide predictive methods for lateralization of mTLE. In this method the classification are represented as a set of rules that are applied sequentially with each rule partitioning an attribute (predictor variable) into a binary response. We performed the CART algorithm 3 to train decision trees using MATLAB r2018a 4. Random Forest method was used for detecting the attributes’ importance. It is an ensemble method, which used a large number of decision trees (e.g. 1000)5.This method applies the randomness in two ways: (i) using a random subset of the observations (bootstrapped sampling) for creating each of the trees, and (ii) each split in a tree is created using a random subset of the candidate variables 5.Results
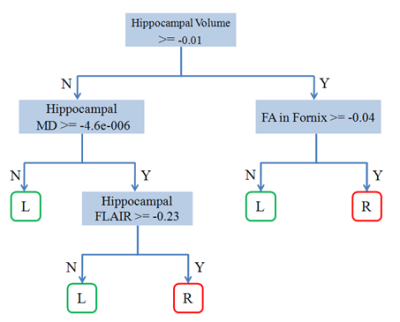
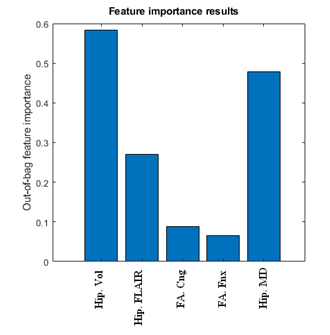
The results of applying the CART model are shown in Fig 1. We used 10-fold cross-validation for training and evaluation trees. Best results with 80% correct rate were selected for establishing the rules for lateralizing left versus right mTLE. decision tree method suggested the hippocampal volume and MD as crucial attributes to predict the laterality, while the FA in cingulum and fornix were not as effective. The result of attribute importance measurement using a random forest of 1000 tree is displayed in Fig.2. As the figure shows, the consented attributes of hippocampal volume and MD were demonstrated as the most influencive attributes. FA in posteroinferior cingulum and FA in fornix crus were shown of the least degree of importance. These results agreed with the results of decision tree reported by some previous papers6,7.Conclusion
The proposed decision tree and random forest methods may provide reliable lateralization of mTLE and introduce an effective tool for a better assessment of efficacy for neuroimaging biomarkers in real clinical setups. The definition of laterality in patients who do not maintain a clear laterality based on clinical and electrophysiological evidence is a potential application of the proposed predictive model.Acknowledgements
We must acknowledge the contribution of Iranian National Brain Mapping Lab (NBNL) and their staffs for MRI data acquisition throughout conducting this project. This work was partially funded and supported by Iran’s National Elites Foundation, National Institute for Medical Research Development (Grant No. 971683), and Cognitive Sciences & Technologies Council (Grant No. 6431), between 2017 and 2019.References
1. Engel J. Mesial Temporal Lobe Epilepsy: What Have We Learned? Neurosci. 2001;7(4):340-352. doi:10.1177/107385840100700410
2. Téllez-Zenteno JF, Hernández-Ronquillo L. A Review of the Epidemiology of Temporal Lobe Epilepsy. Epilepsy Res Treat. 2012;2012:1-5. doi:10.1155/2012/630853
3. Leo Breiman, Jerome Friedman, Charles J. Stone RAO. Classification and Regression Trees. In: Chapman and Hall/CRC. ; 1984:368 Pages.
4. TheMathWorks Inc., Natick, Massachusetts 2015. MATLAB, version 8.5.0.197613 R2015a. 2015. 5. Grömping U. Variable Importance Assessment in Regression: Linear Regression versus Random Forest. Am Stat. 2009;63(4):308-319. doi:10.1198/tast.2009.08199
6. Nazem-Zadeh MR, Elisevich K V., Schwalb JM, Bagher-Ebadian H, Mahmoudi F, Soltanian-Zadeh H. Lateralization of temporal lobe epilepsy by multimodal multinomial hippocampal response-driven models. J Neurol Sci. 2014;347(1-2):107-118. doi:10.1016/j.jns.2014.09.029
7. Mahmoudi F, Elisevich K, Bagher-ebadian H, et al. Data mining MR image features of select structures for lateralization of mesial temporal lobe epilepsy. 2018;i:1-19.