0962
Quantification of Left Ventricular Strain by Joint Analysis of 3D Tagging and Cine MR Images1Institute for Biomedical Engineering, University and ETH Zurich, Zürich, Switzerland, 2Solid Mechanics Laboratory, École Polytechnique, Palaiseau, France
Synopsis
Prognostic value of deformation metrics extracted from cine MR images has been shown by numerous studies in the clinical setting, but with some limitations to detect local changes in the myocardium. Tagged CMR, however, allows tracking material points on the heart wall, revealing local myocardial deformation. Although it is superior to cine MRI in quantification of myocardial torsion, radial strain is often omitted. In this contribution, we enrich our registration technique with the capability to combine cine and tagged in-vivo CMR images, improving accuracy of regional strain assessment from tagged images while acquiring global deformation from cine CMR images.
Purpose
To combine the advantages of two imaging techniques for improved quantification of left ventricular (LV) function: obtaining global shape information from cine Magnetic Resonance (MR) images and temporal evolution of torsion from 3DTAG MRI.Methods
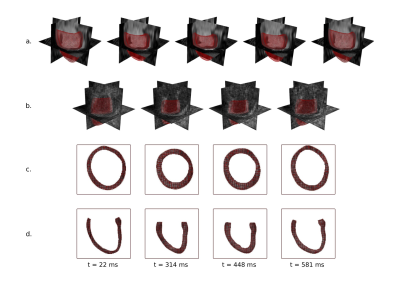
In this study, 17 datasets from a porcine study were used. Imaging was performed on a 1.5T clinical Philips Achieva MR system (32-channel cardiac receiver array). All animal handling procedures and protocols were approved by the Cantonal Veterinary Office (Zurich, Switzerland). The imaging protocol consisted of multi slice cine imaging in short axis view with the following parameters: 1.8×1.8 $$$mm^2$$$ spatial resolution, 8mm slice thickness, 25 heart phases, 1.5ms/3ms TE/TR as well as 3D tagged imaging [2] acquiring three stacks with orthogonal Complementary Spatial Modulation of Magnetization (CSPAMM) [3] tagging using the following imaging parameters: 2×2×5 $$$mm^3$$$ spatial resolution, 110×110×110 $$$mm^3$$$ field-of-view, 26 heart phases, 4mm tagline distance, 4.3ms/9.2ms TE/TR, turbo factor 3, echo planar imaging (EPI) factor 7. The data processing pipeline is illustrated in Figure 1. First, finite element (FE)-based image registration is performed on cine MR images starting from end-systolic (ES) configuration using manually segmented geometries on cine MRI at respective time frames (Figure 1.a). Meshes obtained from cine image registration are consequently translated to 3DTAG image coordinates and displacement fields are interpolated to match the acquisition time (Figure 1.b). 3DTAG images are masked using the obtained meshes at each time frame by keeping the tag line pattern inside the mesh, while the background intensity is assigned to a constant value (Figure 1.c-d). For comparison, FE-based image registration is performed separately on 3DTAG and masked-3DTAG MRI. Performance of masked-3DTAG image registration is assessed in terms of two deformation metrics: Green-Lagrange strains at ES with regard to end-diastole and LV twist. Cine MRI and 3DTAG image registrations are used as ground truths while assessing strain measurements and LV twist, respectively. For all datasets, mean $$$\pm$$$ standard deviations in mid-ventricular radial, circumferential and longitudinal strain components are plotted at ES separately for each image type. LV twist is computed by the difference in averaged rotation through the apical and basal slices at ES, and the mean values at ES are indicated. Strain quantification performance is assessed by computing component-wise strain error at ES for 3DTAG and masked-3DTAG image registrations and reported as mean $$$\pm$$$ standard deviations across hearts. Similarly, mean $$$\pm$$$ standard deviations in LV twist error are computed for Steady State Free Precession (SSFP) cine and masked-3DTAG image analyses.Results
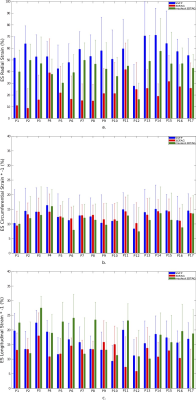
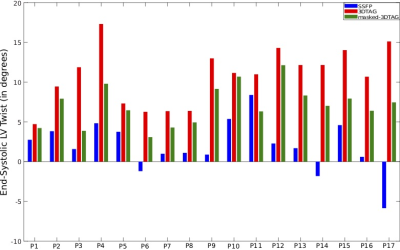
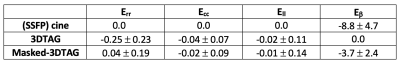
Figure 2 shows mean $$$\pm$$$ standard deviations in mid-ventricular ES radial, circumferential and longitudinal strains acquired from registering 3DTAG and masked-3DTAG images in addition to SSFP images, which were used as ground truth strains for this study. For radial strain quantification, masked-3DTAG image registration performs better compared to 3DTAG images except for data set P12. Circumferential and longitudinal ES strains are not sensitive to different image types. LV twist values (in degrees) at ES are represented in Figure 3. Masked-3DTAG images yield improved LV twist compared to SSFP images. The corresponding values are reported in Table 1. Masked-3DTAG image registration provides a reduced averaged radial strain error from -0.25 $$$\pm$$$ 0.23 for 3DTAG images to 0.04 $$$\pm$$$ 0.19. Error in circumferential and longitudinal strain components are lower, with masked-3DTAG image registration still providing better performance. LV twist error is computed for cine and masked-3DTAG registration. Masked-3DTAG images perform superior to cine image registration in terms of torsion quantification while still performing reasonably well in global motion characterization, decreasing the twist error (in degrees) from -8.8 $$$\pm$$$ 4.7 to -3.7 $$$\pm$$$ 2.4 (Table 1, Equation 8).Discussion $$$\&$$$ Conclusion
Cine MRI and 3DTAG images have their own advantages and disadvantages while quantifying LV deformation. Cine MRI offer higher spatial resolution and hence potentially benefit in particular radial strain calculation, in addition to providing good tracking of the shape. However, tracking algorithms generally fail to detect regional deformation due to lack of contrast through ventricular wall. CMR tagging allows for tracking material points in the myocardium, however, saturation bands generated in the fully tagged images result in partial voluming of blood-myocardial contrast, making it impossible to accurately delineate the endocardial and epicardial borders.This study shows that combined processing of cine MRI and 3DTAG images provides better quantification of LV deformation as either data source alone.
Acknowledgements
The authors acknowledge funding of the Swiss National Science Foundation (SNF) Research Grants Nr. CR23I3_166485 and PZ00P2_174144, as well as of the French National Research Agency (ANR) Research Grant Nr. ANR-10-EQPX-37.References
[1] M. Genet, C. T. Stoeck, C. Von Deuster, L. C. Lee, and S. Kozerke, “Equilibrated Warping: Finite Element Image Registration with Finite Strain Equilibrium Gap Regularization,” Med. Image Anal., vol. 50, pp. 1–22, 2018.
[2] A. K. Rutz, S. Ryf, S. Plein, P. Boesiger, and S. Kozerke, “Accelerated whole-heart 3D CSPAMM for myocardial motion quantification,” Magn. Reson. Med., 2008.
[3] S. E. Fischer, G. C. McKinnon, S. E. Maier, and P. Boesiger, “Improved myocardial tagging contrast,” Magn. Reson. Med., 1993.
Figures