0938
Tumorous Tissue Characterization in Diffuse Glioma Based on 1H-MRS Data Employing 1D Convolutional Neural Networks1Department of Medical Physics and Biomedical Engineering, Tehran university of Medical Science, Tehran, Iran (Islamic Republic of), 2Quantitative MR Imaging and Spectroscopy Group, Research Center for Molecular and Cellular Imaging, Tehran, Iran (Islamic Republic of), 3Center for Biomedical Image Computing and Analytics (CBICA), University of Pennsylvania, Philadelphia, PA, United States, 4Department of Radiology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, United States, 5Biomaterials Engineering, School of Metallurgy and Materials Engineering, Iran University of Science and Technology, Tehran, Iran (Islamic Republic of)
Synopsis
Characterization of intra-tumour subregions in diffuse gliomas helps to guide biopsy procedure and to determine extent of tumour infiltration in the brain tissues. Conventional MRI cannot accurately differentiate intra-tumour subregions, including the most active tumour component and infiltrated edema (IE) from each other and from the normal tissue (NT). In this work, we explore the potential of differentiation of brain tumorous tissue subregions (tumour core, infiltrated edema, normal tissue, bone, and non-brain areas) based on 1H-MRS data using artificial intelligence (AI) techniques.
INTRODUCTION
Diffuse gliomas are characterized with spatial intra-tumour variability within their microenvironment, which is partly responsible for their grim prognosis1. Currently, contrast-enhanced T1-weighted (CE T1-w) and T2-weighted MR imaging are applied for guiding targeted biopsy and surgical/treatment planning procedures for gliomas2,3. Nonetheless, relying upon these images cannot sufficiently stratify tumorous regions including the most active tumour component and infiltrated edema (IE) from each other and from the normal tissue (NT). Recently, deep learning (DL) algorithms have been proposed for quantification of 1H-MRS data and are becoming a novel tool to solve difficult signal processing problem for in-vivo 1H-MRS 4. In this study, we investigate two approaches including: one-dimensional convolutional neural network (1D CNN) and support vector machines (SVM) to explore the role of CSI spectral data for differentiation of biopsy-approved active tumor (AT) tissue representing the tumour core, infiltrated edema (IE) and normal tissue (NT). Moreover, bone and air are included in our study as our additional regions.METHODS
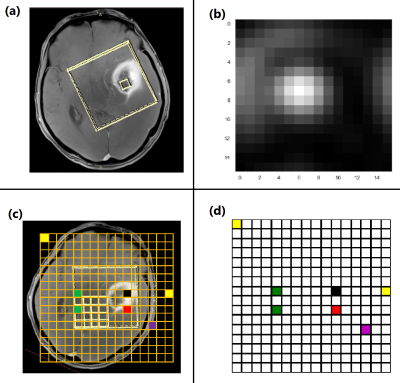
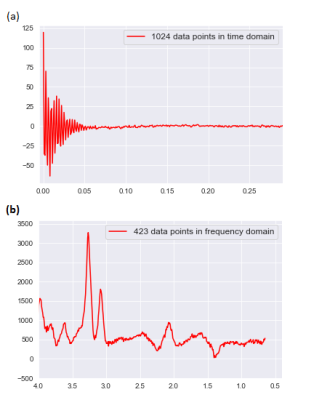
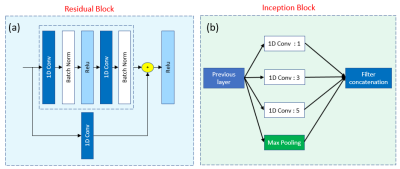
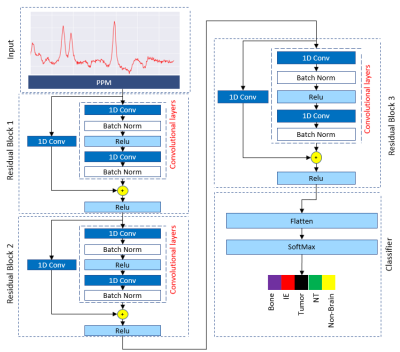
This study was approved by the Institutional Review Board (IRB) and informed consent was obtained. Pre-surgical MRI was performed for eight patients diagnosed with diffuse glioma based on their initial MRI scans5 and on a 3T scanner (Tim Trio, Siemens). The protocol included MPRAGE pre- and post-contrast T1-w, MPRAGE T2-w, T2-FLAIR. Chemical shift imaging acquisition protocol was Point-Resolved Spectroscopy (PRESS) pulse sequence with TE = 135 ms, TR = 1500 ms, 1024 data-points. The 3D box was adapted to the size of tumour. Automatic shimming of magnetic field was performed before data acquisition. T2-w images were selected as reference images to overlay the processed 1H-MRS spectral array. An expert radiologist identified 16x16 rectangular regions-of-interest (ROIs) on CE-T1w MPRAGE images for image-guided needle biopsy sampling. Thirty-four regions were sampled by a neurosurgeon and the specimens were evaluated by a pathologist to be attributed to AT, IE, or NT tissue subregions (7 AT, 8 IE, and 8 NT). The pre-surgical ROIs were overlaid on both the co-registered CSI and images to extract the signal intensity of these labelled ROIs in a color coded grid (Fig. 1) for each patient. Each color in CSI grid (Fig. 1. d) represent a tissue type in CSI. 29 and 126 signals were extracted as bone and out-of-skull (non-brain, air) areas. To avoid “Curse of Dimensionality” we used Fourier transformation (FT) to convert the free-induction decay (FID) signals of 1024 points to particles per million (PPM) scale of 423 frequency points (Fig. 2) this pre-processing can reduce noise and increase SNR. Cross-validated SVM and two 1D CNN classifiers (Fig. 3) were applied to classify samples into AT, IE, NT, Bone, and Air groups. Classification performance was evaluated based on accuracy, sensitivity, and specificity.RESULTS
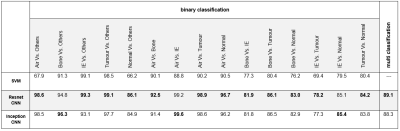
Table1 presents results for tissue characterization using SVM, 1-D Resnet CNN, and 1-D inception CNN. CNNs showed better results for tissue characterization in comparison with SVM. In binary classification, accuracy of SVM, Resnet, and Inception to detect IE were 99.1%, 99.3%, 93.1% respectively and accuracy of above-mentioned algorithms to detect tumor were 98.5%, 99.1%, 97.7%. Over all tissue characterization of our proposed Resnet-based network (Fig. 4) showed better performance with accuracy of 89.1% in comparison with Inception with accuracy of 88.3%.DISCUSSION AND CONCLUSIONS
Lack of sensitive and specific quantitative imaging biomarkers for realizing spatial variations of gliomas leads to inaccurate biopsy sampling, which hinders target-specific diagnosis and therapies. In this study, we investigated the potential of AI methods based on 1H-MRS for differentiating subregions within glioma tumors according to pathologically-validated biopsy specimens. It was shown that DL methods, especially Resnet, could accurately characterize the tissue types in diffuse gliomas.Acknowledgements
No acknowledgement found.References
1- Claes A, Idema AJ, Wesseling P. Diffuse glioma growth: a guerilla war. Acta neuropathologica. 2007; 114(5):443-458.
2- Carlsson SK, Brothers SP, Wahlestedt C. Emerging treatment strategies for glioblastoma multiforme. EMBO molecular medicine. 2014; 6(11):1359-1370.
3- Chamberlain MC. Radiographic patterns of relapse in glioblastoma. Journal of neuro-oncology. 2011; 101(2):319-323.
4- Hatami, N., Sdika, M., & Ratiney, H. Magnetic Resonance Spectroscopy Quantification using Deep Learning. 21st International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2018). 2018
5- Fathi Kazerooni A, Nabil M, Zeinali Zadeh M, et al. Characterization of active and infiltrative tumorous subregions from normal tissue in brain gliomas using multiparametric MRI. Journal of Magnetic Resonance Imaging. 2018;48(4):938-950.
6- Kazerooni AF, Mohseni M, Rezaei S, Bakhshandehpour G, Rad HS. Multi-parametric (ADC/PWI/T2-w) image fusion approach for accurate semi-automatic segmentation of tumorous regions in glioblastoma multiforme. Magnetic Resonance Materials in Physics, Biology and Medicine. 2015; 28(1):13-22.
Figures