0937
Wavelet Oversampling for Imbalance Childhood Brain Tumour Classification
Dadi Zhao1,2, James T. Grist1,2, Heather E.L. Rose1,2, Yu Sun1,2, and Andrew C. Peet1,2
1Institute of Cancer and Genomic Sciences, University of Birmingham, Birmingham, United Kingdom, 2Department of Oncology, Birmingham Children's Hospital, Birmingham, United Kingdom
1Institute of Cancer and Genomic Sciences, University of Birmingham, Birmingham, United Kingdom, 2Department of Oncology, Birmingham Children's Hospital, Birmingham, United Kingdom
Synopsis
Classifying imbalance childhood brain tumours through 1H-MRS metabolite profiles remains a challenging problem. We presented an alternative oversampling method, wavelet oversampling (WvOS). Different from the classic Synthetic Minority Oversampling TEchnique that oversamples the metabolite profiles, WvOS used the wavelet processed 1H-MRS as the oversampled 1H-MRS, followed by quantification and classification. As the result, WvOS can provide dramatically better classification performance than non-oversampled or classic oversampled metabolite profiles. An optimal balanced classification accuracy is achieved as 96% and 72% from 84% and 52% for the 1.5T and 3T cohorts of childhood brain tumours, respectively.
Introduction
Combined with machine learning, metabolite profiles provide support for childhood brain tumour diagnosis1. The classification performance of brain tumours showed promising for common types like medulloblastomas and pilocytic astrocytomas but limited for rare and diverse types like ependymomas. Such imbalance multi-class classification problems could be addressed by oversampling for the minority, which was previously reported by using Synthetic Minority Oversampling TEchnique (SMOTE)2-3. In this abstract, we presented an alternative oversampling method, Wavelet OverSampling (WvOS), which is based on the wavelet decomposition and reconstruction. Compared with SMOTE, WvOS can provide stably better classification performance and could guide further advances of brain tumour classification through deep learning.Theory
The WvOS (Figure 1) is an extension of the Wavelet Noise Suppression (WvNS)4 for proton magnetic resonance spectroscopy. Both WvNS and WvOS assume that some level of noise is presents in the 1H-MRS signals. WvNS generates temporary 1H-MRS signals from the original 1H-MRS signals by using a series of wavelet bases. WvOS selects high-quality 1H-MRS signals from these temporary 1H-MRS signals as the results of oversampling. Specifically, the temporary 1H-MRS signals with the best and the second best signal quality is selected as the noise-suppressed 1H-MRS signal and the oversampled 1H-MRS signal, respectively. If higher oversampling rate is preferred, the temporary 1H-MRS signal with the third best signal quality and the following are used.Methods
Pre-operative patients presenting with a brain tumour like ependymoma, medulloblastoma or pilocytic astrocytoma were recruited in this retrospective study. Clinical 1H-MRS scanning was performed on a 1.5T or 3T scanner in Birmingham Children’s Hospital (TE 30-46ms, TR 1500-2000ms, voxel size 20-80mm3). The raw 1H-MRS were quantified by using TARQUIN (version 4.3.11) with a 1H-brain full basis and screened by two experienced spectroscopists according to the quality of the spectrum. Overall signal-to-noise ratio (SNR) was used to define the quality of 1H-MRS signals. The 1H-MRS analysis and brain tumour classification were conducted separately for the 1.5T and 3T cohorts. Metabolite profiles were used as the features for brain tumour classification. Noise suppression for 1H-MRS was conducted through WvNS with one level of decomposition in the frequency domain4. Metabolites were ranked and selected based on their multi-class area under the curve, and the number of metabolites used for classification was no more than the number of smallest tumour group. Oversampling for the minority was conducted through WvOS and compared to SMOTE. Classification experiments were conducted through linear discriminant analysis and support vector machine with a linear kernel. The classification accuracy was determined through leave-one-out and ten-fold cross validation and compared between non-oversampled and oversampled post-noise suppression 1H-MRS through a Wilcoxon signed-rank test.Results
Totally 82 patients were enrolled to the 1.5T cohort (ependymomas, N=12; medulloblastomas, N=31; pilocytic astrocytomas, N=39), and 20 patients were enrolled to the 3T cohort (ependymomas, N=2; medulloblastomas, N=8; pilocytic astrocytomas, N=10). The classification performance of the oversampled metabolite profiles showed significantly better than the non-oversampled metabolite profiles (P<.05, Figure 2-3), among which WvOS provides significantly better classification performance than non-oversampled or SMOTE-based oversampled metabolite profiles (P<.05). For example, according to linear discriminant analysis and leave-one-out cross validation, WvOS showed the optimal balanced classification accuracy as 96% for the 1.5T cohort (Figure 2) and 72% for the 3T cohort (Figure 3), which is significantly improved from 84% for the 1.5T cohort and 52% for the 3T cohort before oversampling, and 87% for the 1.5T cohort and 58% for the 3T cohort through SMOTE, respectively (P<.05).Discussion
Childhood brain tumour classification through metabolite profiles in clinical diagnosis has met challenges including the limited signal quality, the performance of fitting, the selection of metabolites and the imbalance group size, which has led to limited classification accuracy of childhood brain tumours. Recent progress has shown improved classification accuracy through novel feature selection and noise suppression5, while the issue of limited group size for the rare tumour types like ependymomas remains to be investigated.SMOTE is a useful tool to oversample the minority to reduce the negative affect of imbalance group size3. However, SMOTE can be used for metabolite profiles instead of the 1H-MRS, which makes it vulnerable to the limited fitting performance and could generate unpredictable and inaccurate results. In contrast, the WvOS used in-depth processing on the raw 1H-MRS and guaranteed the reliability of oversampled results.
Due to the presence of noise, the ground-truth of metabolite profiles remain undetectable, thus the data reliability is similar between the noise-suppressed and oversampled 1H-MRS. Due to the difficulty of clinical data collection, deep learning was rarely reported in classifying childhood brain tumours through 1H-MRS. Benefited from the rich variation of wavelet profiles, it is possible to generate a large-scale 1H-MRS dataset from a small clinical cohort. Therefore, the current method has the potential to guide the application of deep learning in classifying childhood brain tumours through 1H-MRS.
Conclusion
Wavelet oversampling provides an alternative solution with better performance for imbalance classification of childhood brain tumour classification.Acknowledgements
We would like to acknowledge funding from the Cancer Research UK and EPSRC Cancer Imaging Programme at the Children’s Cancer and Leukaemia Group (CCLG) in association with the MRC and Department of Health (England) (C7809/A10342), the Cancer Research UK and NIHR Experimental Cancer Medicine Centre Paediatric Network (C8232/A25261), Health Data Research UK (HDR UK) and the Children’s Research Fund.References
- Davies NP, Wilson M, Harris LM et al. Identification and characterisation of childhood cerebellar tumours by in vivo proton MRS. NMR in Biomedicine. 2008;21:908-918.
- Zarinabad N, Wilson M, Gill SK, Manias KA, Davies NP, Peet AC. Multiclass imbalance learning: Improving classification of pediatric brain tumors from magnetic resonance spectroscopy. Magnetic Resonance in Medicine. 2017;77:2114-2124.
- Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: synthetic minority over-sampling technique. Journal of Artificial Intelligence Research. 2002;16:321-357.
- Zhao D, Grist JT, Sun Y, Peet AC. Impact of wavelets and apodisation in magnetic resonance spectroscopy quality for paediatric brain tumours. Proceedings of the Annual Meeting of ISMRM, Montréal, Canada. 27: 4243.
- Zhao D, Grist JT, Sun Y, Sawlani V, Peet AC. Optimised paediatric brain tumor diagnosis by using in vivo magnetic resonance spectroscopy and machine learning. Proceedings of the Annual Meeting of ISMRM, Sydney, Australia 28: 1394.
Figures
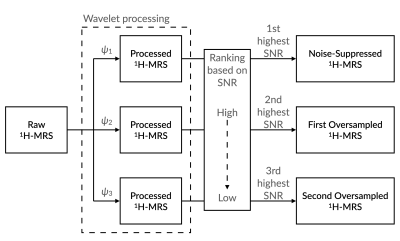
Illustration showing the procedure of generating oversampled proton magnetic resonance spectroscopy (1H-MRS) from the raw 1H-MRS by using Wavelet OverSampling (WvOS). Abbreviations: SNR, signal-to-noise ratios; ψ, wavelet basis.
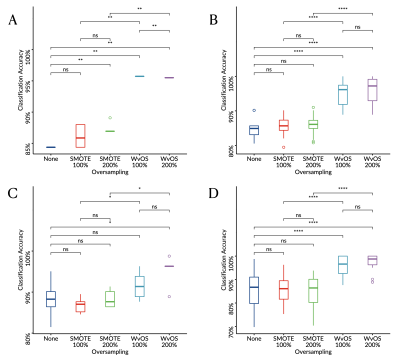
Boxplots showing the balanced classification accuracy for the 1.5T cohort of childhood brain tumours derived through linear discriminant analysis (A, C) or supper vector machine (B, D) with leave-one-out (A-B) or six-fold (C-D) cross validation.
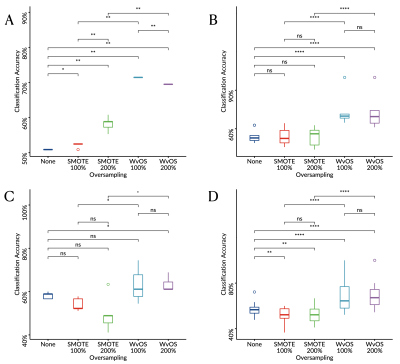
Boxplots showing the balanced classification accuracy for the 3T cohort of childhood brain tumours derived through linear discriminant analysis (A, C) or supper vector machine (B, D) with leave-one-out (A-B) or six-fold (C-D) cross validation.