0935
Estimation of capillary level input function for abbreviated breast Dynamic Contrast-Enhanced MRI using deep learning approach1NYU School of Medicine, New York, NY, United States, 2Center for Advanced Imaging Innovation and Research, New York, NY, United States, 3Center for Biomedical Imaging, NYU, New York, NY, United States, 4Icahn School of Medicine at Mount Sinai, New York, NY, United States, 5Weill Cornell Medicine, New York, NY, United States
Synopsis
This study proposes a deep learning approach to estimate the capillary level input function(CIF) for contrast kinetic model analysis of dynamic contrast enhanced (DCE)-MRI data. Estimation of the CIF for each voxel eliminates the need for the arterial input function and allows automatic end-to-end analysis. We hypothesize that the CIF serves as a more accurate input function that could yield accurate kinetic parameter estimation, which can be used for the diagnosis between the malignant and the benign cancer in the clinical setting. This hypothesis has been tested with a numerical simulation and breast MRI data from an abbreviated exam.
Purpose
Dynamic contrast enhanced(DCE)-MRI has been used as a quantitative tool for diagnosis of breast cancers.1 When analyzing the DCE-MRI data, pharmacokinetic models(PKM), such as the Two compartment exchange model(TCM)2 and the extended-Tofts(eTofts)3, are often used to estimate the pharmacokinetic(PK) parameters that reflect the microenvironment of tissue. All of these model-based analyses method require an input function, generally referred as the arterial input function(AIF). Most studies typically use a global AIF(Ca), either measured at a major artery in the acquired images or from a population-based model. This manual selection of AIF is often very difficult to reproduce, and the variability in the AIF directly results in errors in estimation of PK parameters.4 This study endeavors to develop a deep learning network that could predict the pixel-level capillary input function(CIF)(Cp) to aid PK estimation without using AIF in the setting of abbreviated breast MRI exams of 2.5 min.Methods
Simulation Study:To evaluate the performance of the trained network, we generated 100 patches with randomly selected AIF, simulating either malignant or benign patch. Each voxel in a patch was randomly assigned with the kinetic parameters within the range acquired from the clinical data and simulated with randomly selected AIF. Upon generating this data, the ground truth CIF was calculated and served as the target for training and validation. The gaussian noise with 0.005 variance was mixed, corresponding to approximately 0.5% of 1mMol of final concentration level at 2.5min mark. For each patch, the CIF(Cp) was predicted from the trained network. Then the PKM analysis on the center voxel of each patch was conducted in three different ways: (1) the same AIF(Ca) used in generating the patch with TCM, (2)the population averaged AIF(Ca,pop) with TCM, and (3)the proposed approach with the predicted Cp using eTofts. The blood flow(Fp) was also estimated by fitting predicted Cp into TCM model with the population averaged AIF(Cp+ Ca,pop).
Deep Learning Network:
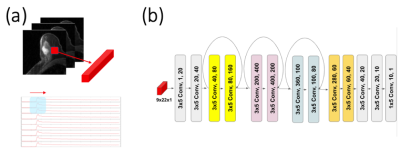
We trained a deep neural network for small patches of dynamic images, assuming the tissue characteristics are similar within a small patch and also all would share a similar capillary level input function. Our network receives a patch of 3-by-3 voxels(n) with multiple frames(t) as the input and predicts the capillary-level input function for the center voxel. To capture features from the changes in temporal domain, we aligned all the pixels in the first dimension and laid out all temporal domain in the second dimension(n x t), as depicted in Figure 1a. We generated the total of one million patches, same way as in the simulation, in which 90% were used in training and 10% were used in validation.
Clinical Study:
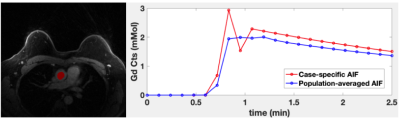
Breast DCE-MRI was performed on the patients with the malignant (n=23) and benign (n=33) lesions. The golden angle radial stack-of-star 3D spoiled GRE sequence was used to acquire 288 spokes for 2.5 min with the contrast injection(gadobutrol, 0.1mM/kg body weight) at 1 min into the scan. All exams were performed on a 3T scanner and the scan parameters include TE/TR = 1.8/4.87ms, flip-angle=10 degrees, matrix size of 320x320x83 and the resolution of 1x1x1.1mm. For the image reconstruction, GRASP-Pro approach5 was used with 13 spokes per frame, yielding ~6.5s per frame temporal resolution. The case-specific AIF was acquired from the aorta in each data set, as shown in Figure 2. The population-averaged AIF was also computed by averaging all AIFs. Each lesion was divided into patches using a sliding window, and the CIF for the center voxel was predicted from the network. We performed the PKM analysis on the “hot-spot” of lesions, defined as the 10% of highest enhancing voxels in a lesion. Similar PKM analysis as in the simulation was conducted. The Receiver Operating Characteristic(ROC) curve analysis was used to assess the diagnostic performance of each method.
Results
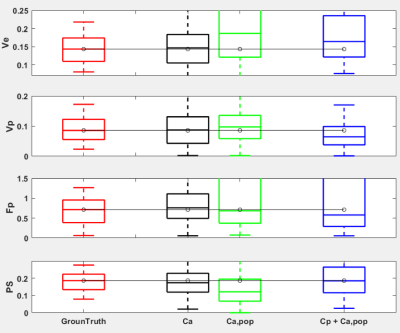
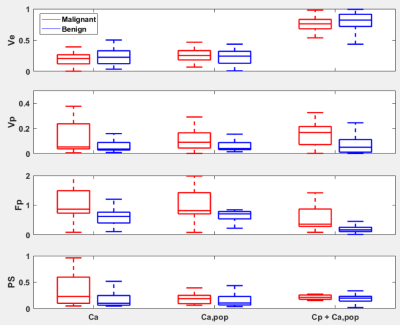
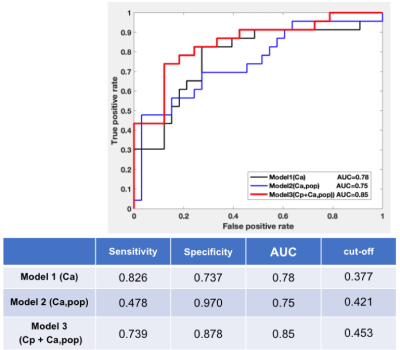
Figure 3 shows the estimated kinetic parameters from the three different approaches. The conventional approach with Ca, maintains the accuracy of the parameter estimation, but when the population-averaged AIF Ca,pop was used instead, the inaccurate estimation with median error of 53% was observed. However, the proposed AI-based approach exhibits better accuracy with the median error of 39%. Figure 4 shows the PKM analysis of the clinical data, where it shows that the PK parameters are in similar ranges among the three methods including the Cp+Ca,pop method which does not involve any manual selection of AIF. Among the estimated PK parameters, Fp was found to have more clear separation between the malignant and the benign lesions. As shown in Figure 5, the Fp from our model yields the best diagnostic performance to distinguish the malignant group from the benign group with AUC=0.85, sensitivity of 74% and specificity of 88%.Discussion and Conclusion
The proposed deep-learning network to estimate the CIF has demonstrated promising performance for accurately estimating PK parameters in breast MRI data with an abbreviated protocol of 2.5min. From the simulation, we observe that the accuracy of PK parameter estimation has maintained in our CIF approach without using the case-specific AIF. In the clinical data, we found that the diagnostic performance was not reduced by using the network estimated CIF, along with the PK parameters in agreement with the literature findings.6Acknowledgements
NIH R01CA219964, R01CA160620, UG3CA228699, R01EB024532, P41EB017183.
References
1. Kim SG, Freed M, Leite APK, Zhang J, Seuss C, Moy L. Separation of benign and malignant breast lesions using dynamic contrast enhanced MRI in a biopsy cohort. Journal of Magnetic Resonance Imaging. 2017;45(5):1385-93.
2. Sourbron SP, Buckley DL. On the scope and interpretation of the Tofts models for DCE‐MRI. Magnetic resonance in medicine. 2011;66(3):735-45.
3. Sourbron S, Buckley DL. Tracer kinetic modelling in MRI: estimating perfusion and capillary permeability. Physics in Medicine & Biology. 2011;57(2):R1.
4. Ziayee F, Müller-Lutz A, Gross J, Quentin M, Ullrich T, Heusch P, et al. Influence of arterial input function (AIF) on quantitative prostate dynamic contrast-enhanced (DCE) MRI and zonal prostate anatomy. Magnetic resonance imaging. 2018;53:28-33.
5. Feng L, Wen Q, Huang C, Tong A, Liu F, Chandarana H. GRASP‐Pro: imProving GRASP DCE‐MRI through self‐calibrating subspace‐modeling and contrast phase automation. Magnetic Resonance in Medicine. 2020;83(1):94-108.
6. Buckley DL. Shutter‐speed dynamic contrast‐enhanced MRI: Is it fit for purpose? Magnetic Resonance in Medicine. 2019;81(2):976-88.
Figures