0864
New insights from the IronTract challenge: Simple post-processing enhances the accuracy of diffusion tractography1Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital and Harvard Medical School, Charlestown, MA, United States, 2University Hospital Center (CHUV) and University of Lausanne (UNIL), Lausanne, Switzerland, 3CIBM Center for BioMedical Imaging, Lausanne, Switzerland, 4École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland, 5Vanderbilt University, Nashville, TN, United States, 6Aalto University School of Science, Espoo, Finland, 7University of Wisconsin, Madison, WI, United States, 8Department of Biomedical Engineering, Eindhoven University of Technology, Eindhoven, Netherlands, 9Department of Radiology and Biomedical Research Imaging Center (BRIC), University of North Carolina, Chapell Hill, NC, United States, 10Department of Neuroscience, Brighton and Sussex Medical School University of Sussex, Brighton, United Kingdom, 11CUBRIC, Cardiff University, Cardiff, United Kingdom, 12NeuroPoly, Polytechnique Montreal, Montreal, QC, Canada, 13Department of Biomedical Engineering, Faculty of Engineering, Yeditepe University, Instanbul, Turkey, 14Neuroscience Research Center, University “Magna Graecia”, Catanzaro, Italy, 15Computational Radiology Laboratory, Boston Children's Hospital, Harvard Medical School, Boston, MA, United States, 16Department of Neurological Surgery, University of Pittsburgh, Pittsburgh, PA, United States, 17Institute of Neurology, University “Magna Graecia”, Catanzaro, Italy, 18Imaging Sciences Institute, University Medical Center Utrecht, Utrecht, Netherlands, 19Neurology Department, Brain Center Rudolf Magnus, University Medical Center Utrecht, Utrecht, Netherlands, 20University of Basel, Basel, Switzerland, 21University of Verona, Verona, Italy, 22Technical University of Denmark, Kongens Lyngby, Denmark, 23Wellesley College, Wellesley, MA, United States, 24DeepHealth, Inc., Cambridge, MA, United States, 25QMENTA, Inc., Barcelona, Spain, 26Department of Pharmacology and Physiology, University of Rochester School of Medicine, Rochester, NY, United States
Synopsis
We present results from round 2 of IronTract, the first challenge to evaluate the accuracy of tractography using i) tracer injections and diffusion MRI from the same macaque brains, and ii) DSI and HCP two-shell diffusion acquisition schemes. In round 1, only two teams achieved similarly high performance between the two different injection sites that we used for training and validation. Here we investigate the extent to which this was due to the pre- and post-processing used by those teams. We show that, when other teams use the same pre- and post-processing, their accuracy and robustness can improve as well.
Introduction
The IronTract challenge evaluates the accuracy of diffusion MRI (dMRI) tractography methods by comparing them to anatomical tracing, using dMRI and tracer data from the same macaque brains. We investigate which dMRI analysis methods lead to optimal accuracy for the two-shell acquisition scheme of the lifespan and disease HCP. Results from round 1 (R1) showed that, when analysis methods were optimized, the HCP acquisition could achieve similar accuracy as a more demanding DSI acquisition1. However, only two out of twelve teams could achieve as high performance on the injection/seed area used for validation (vlPFC) as on the one used for training (frontal pole). Here we seek to disentangle the contributions of the dMRI pre- and post-processing methods from those of the orientation reconstruction and tractography methods, by asking all participants in round 2 (R2) to use the same pre- and post-processing as the two teams that achieved robustness across the two injection sites in R1.Methods
Details on data collection, including in-vivo tracing and ex-vivo dMRI, have been described previously1-3. In R2, all teams used data that had undergone pre-processing by Team1 (denoising4 and Gibbs ringing correction with MRtrix35,6, and motion/eddy-current correction with FSL7,8). All teams were also provided with scripts that implemented the R1 post-processing strategies of Team1 (Gaussian filtering with sigma=0.5 to increase coverage, followed by iterative thresholding of 200 steps on the log of the streamline count) and Team2 (inclusion ROIs from the PennCHOP macaque atlas9, based on general knowledge of projections of the prefrontal cortex). The challenge was administered on the QMENTA platform (qmenta.com/irontract-challenge/). As in R1, participants were blind to the tracer data. Each team submitted tractography volumes thresholded at multiple levels. For each level, the true positive rate (TPR) and false positive rate (FPR) of tractography was computed by voxel-wise comparison to the tracer data. The score was the area under the ROC curve (AUC), for FPRs in [0,0.3]. Thus the maximum AUC was 0.3. For the training case, participants were shown their AUC after uploading their tractography results. They could repeat this up to ten times and fine-tune the free parameters of their methods to optimize their AUC. They then applied the fine-tuned methods to the data of the validation case. Each of the teams that had participated in R1 had to submit results with the orientation reconstruction and tractography methods that they had used in R1, but could also submit results with new methods. We ranked submissions based on overall best AUC score in the validation case.Results
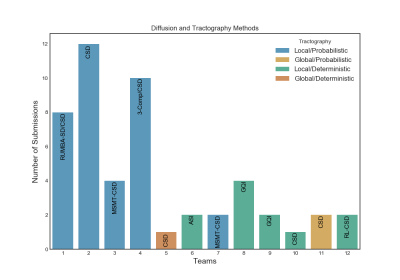
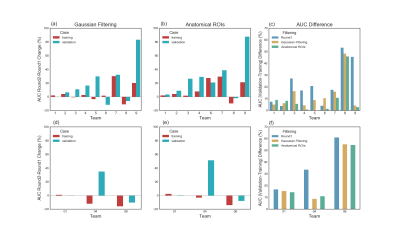
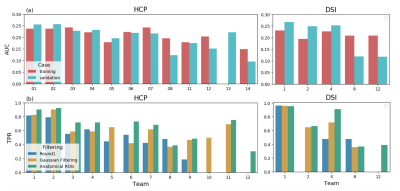
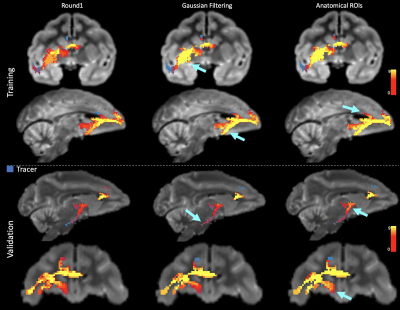
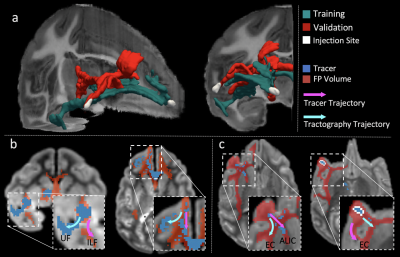
Of the twelve teams that completed R2, nine had also completed R1, while three were new. There was a total of 247 submissions (training: 99, validation: 148) and 50 final submissions that were ranked. Orientation reconstruction and tractography algorithms used by each team are reported in Fig. 1. The performance of most returning teams improved when compared to R1, as a result of applying the pre- and post-processing of Team1 and Team2. This improvement was greater for the validation case (2%-85%) than the training case (2%-30%) (Fig. 2a-b,d-e). For teams that had achieved much lower accuracy on the validation case than the training case in R1, this difference decreased substantially in R2 (Fig. 2c,f). Thus many more teams achieved similar performance between the training and validation case in R2 (Fig. 3a). Of the two post-processing strategies, the use of a priori inclusion ROIs led to consistently higher TPR at the same FPR for all submissions, as expected. Remarkably, the use of Gaussian filtering, which does not assume any prior anatomical knowledge, also improved results for most submissions (Fig. 3b). Only two teams (6 and 8) did not show improvement with Gaussian filtering and one of them (8) did not show improvement with anatomical ROIs. Both of these teams used deterministic tractography, but this was also the case for the team with the greatest % improvement (9). Fig. 4 shows histograms of true positives of tractography compared to the tracing, across all teams. Despite the improved coverage resulting from optimized pre- and post-processing, certain brain regions continue to pose challenges for most teams. Examples of such brain areas where errors occur are shown in Fig. 5.Discussion and Conclusion
The injection sites of the training and validation case, while projecting through similar white-matter pathways (Fig. 5), follow very different routes to reach these pathways9 and pose different challenges to tractography. Most submissions made errors in anatomical locations where fibers from the injection site cross bigger bundles, branch into smaller bundles, travel through bottle-neck regions, or take sharp turns (Fig. 4b-c for some examples). The post-processing used here improved accuracy in these regions and decreased accuracy differences between the two injection sites. However, even after harmonizing pre- and post-processing across teams, Team1 continued to achieve the highest accuracy. When using DSI data, Team1 could reach a TPR as high as 0.96 at FPR=0.1. This suggests that the orientation reconstruction method employed by this team (RUMBA-SD10), in combination with probabilistic tractography, contributed to its high performance.Acknowledgements
Data acquisition was supported by the National Institute of Mental Health (R01-MH045573). Additional research support was provided by the National Institute of Biomedical Imaging and Bioengineering (R01-EB021265). Imaging was carried out at the Athinoula A. Martinos Center for Biomedical Imaging at the Massachusetts General Hospital, using resources provided by the Center for Functional Neuroimaging Technologies, P41-EB015896, a P41 Biotechnology Resource Grant, and instrumentation supported by the NIH Shared Instrumentation Grant Program (S10RR016811, S10RR023401, S10RR019307, and S10RR023043). Andrey Zhylka is supported by the European Union's Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant (765148).The Wisconsin group acknowledges the support from a core grant to the Waisman Center from the National Institute of Child Health and Human Development (IDDRC U54 HD090256).References
1. Lehman JF, Greenberg BD, McIntyre CC, Rasmussen SA, Haber SN. Rules ventral prefrontal cortical axons use to reach their targets: implications for diffusion tensor imaging tractography and deep brain stimulation for psychiatric illness. Journal of neuroscience. 2011;31:10392–10402.
2. Safadi Z, Grisot G, Jbabdi S, Behrens TE, Heilbronner SR, McLaughlin NCR, Mandeville J, Versace A, Phillips ML, Lehman JF, Yendiki A, Haber SN. Functional Segmentation of the Anterior Limb of the Internal Capsule: Linking White Matter Abnormalities to Specific Connections. J Neurosci. 2018 Feb 21;38(8):2106-2117.
3. Maffei, C., G. Girard, K. G. Schilling, N. Adluru, D. B. Aydogan, A. Hamamci, F.-C. Yeh, M. Mancini, Y. Wu, A. Sarica, A. Teillac, S. H. Baete, D. Karimi, Y.-C. Lin, F. Boada, N. Richard, B. Hiba, A. Quattrone, Y. Hong, D. Shen, P.-T. Yap, T. Boshkovski, J. S. W. Campbell, N. Stikov, G. B. Pike, B. B. Bendlin, A. L. Alexander, V. Prabhakaran, A. Anderson, B. A. Landman, E. J. Z. Canales-Rodríguez, M. Barakovic, J. Rafael-Patino, T. Yu, G. Rensonnet, S. Schiavi, A. Daducci, M. Pizzolato, E. Fischi-Gomez, J.-P. Thiran, G. Dai, G. Grisot, N. Lazovski, A. Puente, M. Rowe, I. Sanchez, V. Prchkovska, R. Jones, J. Lehman, S. Haber and A. Yendiki (2020). The IronTract challenge: Validation and optimal tractography methods for the HCP diffusion acquisition scheme. ISMRM (oral presentation).
4. Veraart, J.; Novikov, D.S.; Christiaens, D.; Ades-aron, B.; Sijbers, J. & Fieremans, E. Denoising of diffusion MRI using random matrix theory. NeuroImage, 2016, 142, 394-406, doi: 10.1016/j.neuroimage.2016.08.016
5. Tournier JD., Calamante, F, Connelly, A. MRtrix: Diffusion tractography in crossing fiber regions. Int. J. Imaging Syst. Technol. 22, 53–66 (2012). DOI 10.1002/ima.22005.
6. Kellner, E; Dhital, B; Kiselev, V.G & Reisert, M. Gibbs-ringing artifact removal based on local subvoxel-shifts. Magnetic Resonance in Medicine, 2016, 76, 1574–1581.
7. Jesper L. R. Andersson and Stamatios N. Sotiropoulos. An integrated approach to correction for off-resonance effects and subject movement in diffusion MR imaging. NeuroImage, 125:1063-1078, 2016.
8. Andersson JL, Skare S, Ashburner J (2003) How to correct susceptibility distortions in spin-echo echo-planar images: application to diffusion tensor imaging. Neuroimage 20: 870–888
9. Feng L, Jeon T, Yu Q, Ouyang M, Peng Q, Mishra V, Pletikos M, Sestan N, Miller MI, Mori S, Hsiao S, Liu S, Huang H, 2017. Population-averaged macaque brain atlas with high-resolution ex vivo DTI integrated into in vivo space. Brain Struct Funct. 222(9): 4131-4147.
10. Canales-Rodríguez EJ, Daducci A, Sotiropoulos S, Caruyer E, Aja-Fernández S, Radua J, Yurramendi Mendizabal JM, Iturria-Medina Y, Melie- García L, Alemán-Gómez Y, Thiran JP, Sarró S, Pomarol-Clotet E, Salvador R. Spherical Deconvolution of Multichannel Diffusion MRI Data with Non-Gaussian Noise Models and Spatial Regularization. PLoS One. 2015;10(10):e0138910.
11. Tournier JD, Calamante F, Connelly A. Robust determination of the fibre orientation distribution in diffusion MRI: non-negativity constrained superresolved spherical deconvolution. Neuroimage. 2007;35(4):1459-1472.
12. Jeurissen B, Tournier JD, Dhollander T, Connelly A, Sijbers J. Multi-tissue constrained spherical deconvolution for improved analysis of multi-shell diffusion mri data. NeuroImage. 2014;103:411-426, 2014.
13. Dhollander T, Mito R, Raffelt D, Connelly A., \u201cImproved white matter response function estimation for 3-tissue constrained spherical deconvolution\u201d, ISMRM 2019, Montreal.
14. Tran G and Yonggang Shi. "Fiber orientation and compartment parameter estimation from multi-shell diffusion imaging." IEEE transactions on medical imaging. 2015;34(11):2320-2332.
15. Wu, Y. et al. Asymmetry spectrum imaging for baby diffusion tractography. In International Conference on Information Processing in Medical Imaging. 319–331 (Springer, 2019).
16. Yeh FC, Wedeen VJ, Tseng WY. Generalized q-sampling imaging. IEEE TMI. 2010;29(9)5.
17. Guo, F., Leemans, A., Viergever, M. A., Dell’Acqua, F., & De Luca, A. (2020).Generalized Richardson-Lucy (GRL) for analyzing multi-shell diffusion MRI data.NeuroImage, 218(April), 116948. https://doi.org/10.1016/j.neuroimag
Figures