0833
B-spline Parameterized Joint Optimization of Reconstruction and K-space Sampling Patterns (BJORK) for Accelerated 2D Acquisition1Biomedical Engineering, University of Michigan, Ann Arbor, MI, United States, 2Electrical Engineering and Computer Science, University of Michigan, Ann Arbor, MI, United States
Synopsis
The proposed approach, BJORK, provides a robust and generalizable workflow to jointly optimize non-Cartesian sampling patterns and a physics-informed reconstruction. Several approaches, including re-parameterization of trajectories, multi-level optimization, and non-Cartesian unrolled neural networks, are introduced to improve training effect and avoid sub-optimal local minima. The in-vivo experiments show that the networks and trajectories learned on simulation dataset are transferable to the real acquisition even with different parameter-weighted MRI contrasts and noise-levels, and demonstrate improved image quality compared with previous learning-based and model-based trajectory optimization methods.
Introduction
The work proposes an approach for jointly optimizing the reconstruction algorithm and sampling trajectories with regard to reconstruction quality. The trajectories are parameterized with quadratic B-spline kernels to reduce the number of parameters and enable multi-scale optimization, helping to avoid sub-optimal local minima. An efficient non-Cartesian unrolled neural network reconstruction and an accurate approximation for backpropagation through NUFFT operator are combined to accurately reconstruct multi-coil, undersampled, non-Cartesian data. To correct the potential eddy-current effect brought by the trajectory, we used a pencil-beam trajectory mapping technique. In both simulation and in-vivo experiments, the learned trajectory demonstrates significantly improved image quality compared to previous trajectory optimization methods under high acceleration ratio.Methods
Problem formulation: Figure 1 shows the overall workflow. The goal of the proposed supervised approach is to use a set of fully sampled training images to optimize jointly a multi-shot k-space sampling pattern $$$\boldsymbol{\omega}$$$ and the parameters $$$\boldsymbol{\theta}$$$ of an unrolled neural network (UNN) reconstruction method by minimizing the following training losses: $$\underset{\omega \in \mathbb{R}^{N_{s} \times 2}, \theta \in \mathbb{R}^{M}}{\operatorname{arg\,min}} \mathbb{E}_{\boldsymbol{x}}\left\|f_{\boldsymbol{\theta}}(\boldsymbol{\omega} ; \boldsymbol{A}(\boldsymbol{\omega}) \boldsymbol{x}+\boldsymbol{\varepsilon})-\boldsymbol{x}\right\| +\phi_{\gamma \Delta t g_{\max}}\left(\left|\boldsymbol{D}_{1} \boldsymbol{\omega}\right|\right)+\phi_{\gamma \Delta t^{2} s_{\max }}\left(\left|\boldsymbol{D}_{2} \boldsymbol{\omega}\right|\right),$$where $$$\boldsymbol{x}$$$ is the fully-sampled reference image from the training data set and $$$\boldsymbol{\varepsilon}$$$ is the simulated additive noise. $$$\boldsymbol{D}_{1}$$$ and $$$\boldsymbol{D}_{2}$$$ are the first-order and second-order differential matrices. The second and third terms are the penalties on maximum gradient strength ($$$g_{\max}$$$) and slew rate ($$$s_{\max}$$$). $$$f_{\boldsymbol{\theta}}(\boldsymbol{\omega}; \cdot)$$$ is the UNN reconstruction algorithm for non-Cartesian data [1], with parameters $$$\boldsymbol{\theta}$$$.We parameterize the sampling pattern with 2nd order quadratic B-spline kernels $$$\boldsymbol{\omega}=\boldsymbol{B} \boldsymbol{c}$$$, where $$$\boldsymbol{B}$$$ is the $$$N_{s}\times L$$$ interpolation matrix and $$$\boldsymbol{c}$$$ is the coefficient. This approach reduces the number of gradient constraints from $$$8N_{s}$$$ to $$$8L$$$ [2]. Also, the parameterization allows multi-level optimization with variable B-spline kernel lengths from large to small.
Joint learning of trajectories and reconstruction: We use the fastMRI initiative dataset [3] to train the proposed model. The initialization of $$$\boldsymbol{\omega}$$$ uses common trajectories, such as radial spokes and spiral interleaves.
Eddy current correction: We used the ‘k-space mapping’ method by exciting a pencil-beam region multiple times using orthogonal slice selection with a 90$$$^{\circ}$$$ and 180$$$^{\circ}$$$ pulse pair [4,5]. Zeroth eddy current phase fluctuation is also subtracted from the acquisition.
Experiments: We compared our method with SPARKLING [6], a model-based trajectory optimization method. For the simulation experiment, a test case consisting of 950 slices were undersampled and then reconstructed w.r.t. different trajectories. The unrolled neural networks (UNN) are fine-tuned for different trajectories. The compressed sensing reconstruction is implemented using the SigPy package [7].
For the in-vivo acquisition, we compare different readout waveforms on a gradient-echo sequence with T1w contrast, and the protocols are detailed in Fig 5(a). Both SPARKLING and BJORK are initialized with the same 20$$$\times$$$ undersampled radial trajectory. The sequences were programmed via TOPPE [5] and implemented on a GE MR750 3.0T scanner with a Nova Medical 32RX head coil. Subjects gave informed consent under local IRB approval.
Results
Figure 2(a) shows the evolution of the learned trajectories. Different widths of the B-spline kernels introduce different levels of improvement as the acquisition is optimized. Also, the results of multi-level optimization and un-parameterized (PILOT-like [8]) were compared. Directly optimizing sampling points introduce only small perturbations of the initialization. Figure 2(b) shows the training set's influence on the learned sampling pattern. Different anatomical features may lead to different spatial encoding strategies, suggesting that scan-specific trajectories may improve imaging quality.Figure 3 exhibits the eddy-current correction results for both BJORK and SPARKLING. Figure 4 showcases the results of simulation experiments. For both learning-based and model-based reconstruction, the BJORK trajectory shows significant improvement compared to un-optimized trajectories and SPARKLING-like trajectories. Figure 5(b) displays the results of the in-vivo study. BJORK has the lowest level of artifacts compared to SPARKLING and undersampled radial trajectories.
Conclusion
This work proposes an efficient learning-based framework for MRI trajectory design. We introduced several methods to improve the training effect, including trajectory parameterization, multi-level training, and accurate approximation of NuFFT’s Jacobian [9]. Simulation and in-vivo results show that these approaches stabilize the training and may help avoid sub-optimal local minima. Further study will investigate sequences with longer readout time, and extending the model to 3D acquisition.Acknowledgements
This work is supported in part by NIH Grants R01 EB023618 and U01 EB026977, and NSF Grant IIS 1838179.References
[1] Aggarwal HK, Mani MP, Jacob M. MoDL: Model-based deep learning architecture for inverse problems. IEEE transactions on medical imaging. 2018 Aug 13;38(2):394-405.
[2] Sun H, Fessler JA, Noll DC, Nielsen JF. Joint design of excitation k-space trajectory and RF pulse for small-tip 3D tailored excitation in MRI. IEEE transactions on medical imaging. 2015 Sep 15;35(2):468-79.
[3] Zbontar J, Knoll F, Sriram A, Murrell T, Huang Z, Muckley MJ, Defazio A, Stern R, Johnson P, Bruno M, Parente M. fastMRI: An open dataset and benchmarks for accelerated MRI. arXiv preprint arXiv:1811.08839. 2018 Nov 21.
[4] Robison RK, Li Z, Wang D, Ooi MB, Pipe JG. Correction of B0 eddy current effects in spiral MRI. Magnetic resonance in medicine. 2019 Apr;81(4):2501-13.
[5] Nielsen JF, Noll DC. TOPPE: A framework for rapid prototyping of MR pulse sequences. Magnetic resonance in medicine. 2018 Jun;79(6):3128-34.
[6] Lazarus C, Weiss P, Chauffert N, Mauconduit F, El Gueddari L, Destrieux C, Zemmoura I, Vignaud A, Ciuciu P. SPARKLING: variable‐density k‐space filling curves for accelerated T2*‐weighted MRI. Magnetic resonance in medicine. 2019 Jun;81(6):3643-61.
[7] Ong F, Lustig M. SigPy: a python package for high performance iterative reconstruction. InProceedings of the ISMRM 27th Annual Meeting, Montreal, Quebec, Canada 2019 May (Vol. 4819).
[8] Weiss T, Senouf O, Vedula S, Michailovich O, Zibulevsky M, Bronstein A. PILOT: Physics-informed learned optimal trajectories for accelerated MRI. arXiv preprint arXiv:1909.05773. 2019 Sep.
[9] Wang G, Noll DC, Fessler JA, Efficient NUFFT Backpropagation for Stochastic Sampling Optimization in MRI. Submitted to ISMRM 2021.
Figures
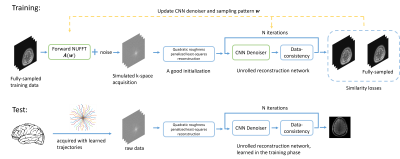
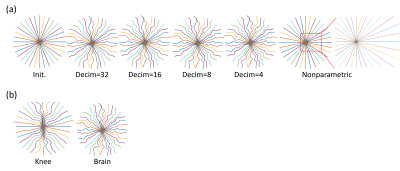
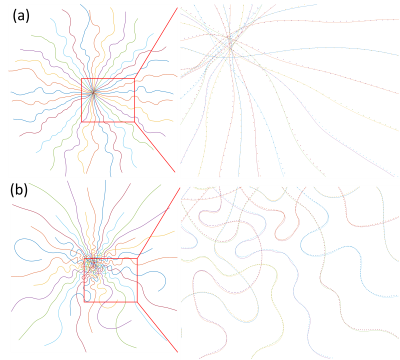
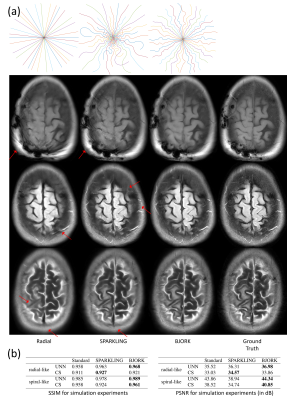
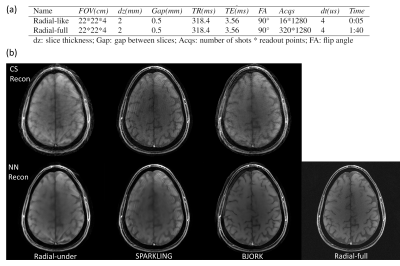
Figure 5. (a) displays the scanning protocols. Radial-like corresponds to the undersampled radial trajectory, SPARKLING trajectory initialized with the undersampled radial trajectory, and BJORK trajectory initialized with the undersampled radial trajectory. (b) showcases one example from the in-vivo experiment. The first row is the CS-based reconstruction and the second row is the unrolled neural network-based reconstruction. For fully-sampled data, conjugate phase reconstruction is displayed.