0820
Prostate Lesion Segmentation on VERDICT-MRI Driven by Unsupervised Domain Adaptation1Centre of Medical Image Computing, University College London, London, United Kingdom, 2Department of Computer Science, University College London, London, United Kingdom, 3Department of Radiology, UCLH NHS Foundation Trust, University College London, London, United Kingdom, 4Division of Surgery & Interventional Science, University College London, London, United Kingdom, 5Centre for Medical Imaging, Division of Medicine, University College London, London, United Kingdom
Synopsis
In this work we utilize unsupervised domain adaptation for prostate lesion segmentation on VERDICT-MRI. Specifically, we use an image-to-image translation method to translate multiparametric-MRI data to the style of VERDICT-MRI. Given a successful translation we use the synthesized data to train a model for lesion segmentation on VERDICT-MRI. Our results show that this approach performs well on VERDICT-MRI despite the fact that it does not exploit any manual annotations.
Introduction
The successful adoption of novel medical imaging modalities for improved automated diagnosis can be delayed by the need of labelled training data. Domain adaptation methods alleviate this problem by exploiting labelled data from a related source domain to train models that generalize well in the target domain despite the absence of annotations for it. In this work we tackle this problem for prostate lesion segmentation and an advanced magnetic resonance imaging (MRI) technique called VERDICT-MRI [1, 2]. VERDICT-MRI is a non-invasive imaging technique that uses a mathematical model to link the diffusion-weighed (DW) signal to microscopic features of tissues. Compared to the naive DW-MRI from multiparametric(mp)-MRI acquisitions, VERDICT-MRI has a richer acquisition protocol to probe the underlying microstructure and reveal changes in tissue features similar to histology. VERDICT-MRI has shown promising results in clinical settings, discriminating normal from malignant tissue [2] and identifying specific Gleason grades [3]. However, the lack of labelled training data does not allow us to use data-driven approaches which could directly exploit the raw VERDICT-MRI. In this work we study the feasibility of using labelled mp-MRI data to train a segmentation network that performs well on VERDICT-MRI despite the lack of annotations which is quite common hurdle for novel imaging techniques.Method
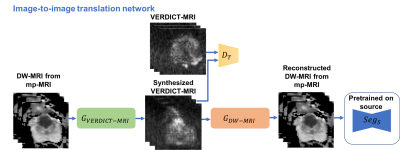
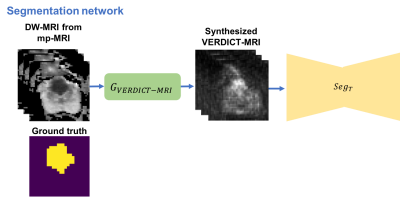
In this work we utilize a pixel-level domain adaptation to train a network for lesion segmentation on VERDICT-MRI by using annotated mp-MRI data. Specifically, we use an image-to-image translation method called MUNIT [5] to synthesize VERDICT-MRI data from annotated mp-MRI data (Fig. 1). During the translation it is important to preserve the lesion on the synthesized VERDICT-MRI since we want to use them along with the corresponding segmentation masks to train the segmentation network. To this end we use a segmentation-based loss in the source domain. GAN-type losses are used to generate realistic VERDICT-MRI data from DW-MRI form mp-MRI acquisitions while standard cycle-consistency losses allow us to preserve the content during the translation. To ensure that the lesions are preserved we use a cyclic-segmentation loss on the cycle-reconstructed images obtained using a pre-trained source-domain network, $$$Seg_S$$$ (Fig. 1). Given a successful translation, we utilize the synthesized data to train a segmentation network for lesion segmentation on VERDICT-MRI (Fig. 2). The segmentation network is trained using only synthesized VERDICT-MRI and the corresponding segmentation masks, without using any real VERDICT-MRI data.Datasets
VERDICT-MRIWe use DW VERDICT-MRI data from 60 men. We acquire VERDICT-MRI images with pulsed-gradient spin-echo sequence (PGSE) using an optimised imaging protocol for VERDICT prostate characterisation with 5 b-values (90-3000 $$$\rm s/ \rm mm^2$$$ ), in 3 orthogonal directions, on a 3T scanner [6]. A separate b $$$= 0$$$ $$$ \rm s/ \rm mm^2$$$ was also acquired before each b-value acquisition. VERDICT-MRI data was registered using rigid registration [7]. A dedicated radiologist contoured the lesions (Likert score of 3 and higher) on VERDICT MRI using mp-MRI for guidance.
DW-MRI from mp-MRI acquisitions.
We use DW-MRI data from the ProstateX challenge dataset consisting of training mp-MRI data acquired from 204 patients. The DW-MRI data were acquired with a single-shot echo planar imaging sequence with diffusion encoding gradients in three directions. Three b-values were acquired (50, 400, 800 $$$\rm s/ \rm mm^2$$$ ) and subsequently, the ADC map and a b-value image at 1400 $$$\rm s/ \rm mm^2$$$ were calculated by the scanner software. Since the ProstateX dataset provides only the position of the lesion, a radiologist manually annotated the lesions on the ADC map using as reference the provided position of the lesion.
Results
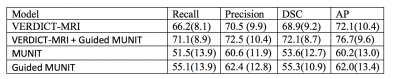
We report results for the following experimentsi) VERDICT-MRI: Fully supervised training on VERDICT-MRI.
ii) VERDICT-MRI + Guided MUNIT: Fully supervised training on real VERDICT-MRI and the synthesized VERDICT-MRI obtained from the image-to-image translation network which is trained using segmentation supervision.
iii) MUNIT: we use MUNIT to map from mp-MRI to VERDICT-MRI without segmentation supervision and then train the segmentation network using the synthesized data.
iv) Guided MUNIT: we use MUNIT and segmentation supervision on the reconstructed mp-MRI images to perform the translation. Then, we use the synthesized images to train the segmentation network.
We employ a nested 5-fold cross validation to select the hyper-parameters and to evaluate the performance. We report the average performance across the outer 5-fold cross validation. We evaluate the performance based on the average recall, precision, dice similarity coefficient (DSC), and average precision (AP). We report the results in Figure 3.
The results show that we can achieve acceptable performance on VERDICT-MRI by training a segmentation model using only synthesized VERDICT-MRI obtained by DW-MRI from mp-MRI acquisitions. We also show that the performance increases when we use segmentation supervision to guide the translation (Guided MUNIT) since the lesion are better preserved in the synthesized images.
Conclusion
In this work we utilize unsupervised domain adaptation to train a model for lesion segmentation on VERDICT-MRI by using labelled DW-MRI from mp-MRI acquisitions. The results show that using this approach we obtain segmentation performance which is close to the one obtained when we train using full supervision on VERDICT-MRI. We report results on VERDICT-MRI but this approach can generalize to other application where there is lack of annotations for the modality of interest.Acknowledgements
This research is funded by EPSRC grand EP/N021967/1. We gratefully acknowledge the support of NVIDIA Corporation with the donation of the GPU used for this research.References
1. Panagiotaki, E., et al.: Noninvasive quantification of solid tumor microstructure using VERDICT MRI. Cancer Research 74, 7, 2014
2. Panagiotaki, E., et al.: Microstructural characterization of normal and malignant human prostate tissue with vascular, extracellular, and restricted diffusion for cytometry in tumours magnetic resonance imaging. Investigative Radiology 50, 4, 2015
3. Johnston, W. E., et al.: INNOVATE: A prospective cohort study combining serum and urinary biomarkers with novel diffusion-weighted magnetic resonance imaging for the prediction and characterization of prostate cancer. BMC Cancer, 16, 816, 2016
4. Johnston, W. E., et al.: Verdict MRI for prostate cancer: Intracellular volume fraction versus apparent diffusion coefficient. Radiology, 291, 2, 2019
5. Huang, X., et al: Multimodal unsupervised image-to- image translation, ECCV, 2018
6. Panagiotaki, E., et al.: Optimised VERDICT MRI protocol for prostate cancer characterisation. In: ISMRM, 2015
7. Ourselin, S., et al.: Reconstructing a 3D structure from serial histological sections. IVC 19, 1, 2001
Figures