0807
The Effect of Activation Functions and Loss Functions on Deep Learning Based Fully Automated Knee Joint Segmentation1Department of Biomedical Engineering, Cleveland Clinic, Cleveland, OH, United States
Synopsis
Studies on systematic evaluations of effects of activation functions and loss functions on deep learning-based automated knee compartments segmentation models are limited. In this work, we present a 2D-UNet model for simultaneous automated bone and cartilage segmentation, and analyze the effect of different activation functions (rectified linear unit[relu], sigmoid and softmax) at all or last layer, and different loss functions (categorical cross-entropy, multiclass dice coefficient loss) with and without surface distance weights, on model performance. The results showed significant performance differences in average surface distance (ASD) between different activation functions. Adding surface distance to loss functions improved segmentation performances.
Introduction
Osteoarthritis (OA) is the most common form of arthritis that affects over 32.5 million U.S. adults1. Quantitative MRI have been proposed as potential imaging biomarkers for early diagnosis, evaluating therapy response and monitoring of OA progression. However, the transition of quantitative MRI in clinical settings requires fast, accurate, fully automatic tissue segmentation to reduce manual segmentation efforts. Although recent works had used deep learning models for fully automatic segmentation of different knee compartments2,3,4, studies on systematic evaluations of effects of activation functions and loss functions on model performance are few. In this work, we present a fully automatic deep learning based segmentation algorithm developed with data from the Osteoarthritis Initiative (OAI) and analyzed the effect of the different activation functions and loss functions on model performance.Methods
507 knee images from the Osteoarthritis Initiative (OAI) data set (relevant scan parameters: FOV=14 cm, Matrix=384×307 zero-filled to 384×384, TE/TR=5/16ms, 160 slices with a thickness of 0.7mm) with manual segmentation for bone (femur bone, tibia bone) and cartilage (femur cartilage, tibial cartilage) were used5. The 507 images were randomly split into 70:20:10 ratio for training, validation, and testing. Deep learning architecture based on 2D U-Net was used to generate segmentation as it had performed well in biomedical image segmentation. The 2D U-Net had a depth of 7 and 14 convolution layers of encoder and decoder. The model consists of 23 millions trainable parameters. The U-Net took 2D MR image slices as input and segmentation masks with five channels (background and four segmentation labels) as reference output. The model provided pixel-wise predicted masks for the knee joint. The Adam optimizer was used with an initial learning rate of 10e-4. Batch size was 10. In training, the input MRI volumes were augmented by random flip along X-axis during runtime. The model was implemented in Python using Keras 2.4.0 and Tensorflow 2.3.0 framework and trained on Google Cloud with NVIDIA Tesla P100 and V100 GPU. The model was trained separately with no activation and three different activation functions (relu, sigmoid, and softmax). Each model was trained for 40 epochs using either categorical cross entropy loss function or multiclass dice coefficient loss function. For each loss function, we tested without and with surface distance loss, with the latter giving more importance to losses around the boundary regions for each output compartments. The segmentation performance was evaluated using (a) Dice coefficient (range between 0-1), which provides overlapping of the automatic segmentation labels and the manual segmentation labels; and (b) Surface distance, which assesses how closely the surfaces between the two segmentations align.Results
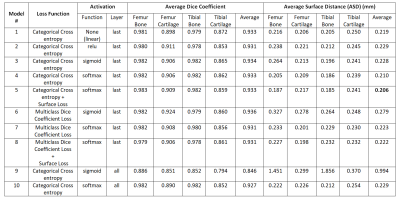
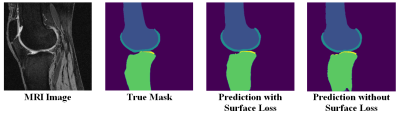
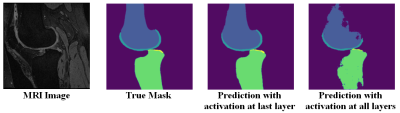
The semantic segmentation performance in terms of average dice coefficients and surface distances for different activation and loss functions on the held-out test set of 51 subjects are listed in Table 1. The model had the lowest ASD when it was trained using categorical cross-entropy loss with surface distance loss (Model #5). Adding surface distance weights to dice and cross entropy loss function improved segmentation performance in terms of ASD for both losses. Fig 1 shows the segmentation improvement due to surface distance weighting in case of multiclass dice loss. Performance with softmax activation at last layer was better than the sigmoid in terms of ASD in both losses. Specifying activation at all layers using sigmoid or softmax degraded performance significantly. In Fig 2, it shows the effect of specifying activation at last layer vs. all layers.Discussion
The models having softmax or sigmoid activations at each up-sampling or down-sampling layers of the UNet (Model #9 and #10) had worst performance as it deactivates some paths during training of the network. All other models showed good bone and cartilage segmentation performance regardless of loss or activation functions. The mean dice coefficient over Model #1 to #8 of bone is higher than cartilage (0.98 vs 0.88), while the ASD were comparable (0.225 vs. 0.229). The reason for differences in dice coefficients between cartilage and bone is due to the smaller cartilage areas compared to bone and we used loss functions with same class’ weights. These results suggest ASD may serve as a metric for segmentation evaluation which is independent to the size of tissue of interest. In addition, the dice coefficient varied little for different activations, but the models with softmax activation at last layer performed better than the sigmoid in terms of ASD because the output segmentation labels were mutually exclusive. Lastly, our results suggest that adding surface distance to the loss function improved the segmentation results in terms of ASD as we give more importance of the losses around the boundary regions of the compartments.Conclusion
In this study, we presented a detailed analysis of activation function and loss function in deep learning-based model to automatically segment knee joints. The model performance was good for a large amount of test data. In future work, we will explore the effect of additional losses such as focal loss, shape aware loss etc. and activation functions such as leaky relu. These analyses may help to build accurate segmentation model which is desired in OA studies based on quantitative MRI.Acknowledgements
No acknowledgement found.References
1. Y. Zhang et al., "Epidemiology of osteoarthritis", Clinics in geriatric medicine, pp. 355-69, 2010
2. Norman B, Pedoia V, Majumdar S. Use of 2D U-Net Convolutional Neural Networks for Automated Cartilage and Meniscus Segmentation of Knee MR Imaging Data to Determine Relaxometry and Morphometry. Radiology. 2018;288(1):177-85.
3. Zhou Z, Zhao G, Kijowski R, Liu F. Deep convolutional neural network for segmentation of knee joint anatomy. MagnReson Med. 2018
4. Gaj, Sibaji, et al. "Automated cartilage and meniscus segmentation of knee MRI with conditional generative adversarial networks." Magnetic Resonance in Medicine 84.1 (2020): 437-449.
5. OAI ZIB :https://amira.zib.de/download.html
Figures