0803
Deep Learning-based Semi-supervised Meniscus Segmentation with Uncertainty Estimation1AI in Radiology Laboratory, Department of Imaging and Interventioanl Radiology, The Chinese University of Hong Kong, Hongkong, Hong Kong
Synopsis
Accurate segmentation of the meniscus is valuable for clinical diagnosis and treatment of knee joint diseases. Due to expensive and time-consuming medical image data annotation, it is challenging to obtain sufficient labeled data for deep learning-based segmentation of meniscus. We investigated deep-learning based semi-supervised approaches with uncertainty estimation for meniscus segmentation using MR images.
Introduction
Automatic meniscus segmentation using magnetic resonance (MR) images is valuable for diagnosis and prognosis of knee joint diseases. Recent developments in deep learning-based meniscal segmentation models have heightened the demand for annotated data. However, it is challenging to accurately annotate large amount of meniscal images manually by experts in the field. It is highly desirable to develop deep learning-based meniscus segmentation methods which can utilize substantial unlabeled data resources. Uncertainty estimation is a potential direction that can be leveraged to generate a pseudo label for corresponding unannotated input. In this work, we evaluated the dropout-based Bayesian neural network and demonstrated it is capable of efficiently utilizing additional unlabeled data for better performance of meniscus segmentation. 1 Besides, we explored three different uncertainty measures in meniscus segmentation results and demonstrated what features these measures can capture. 2Methods
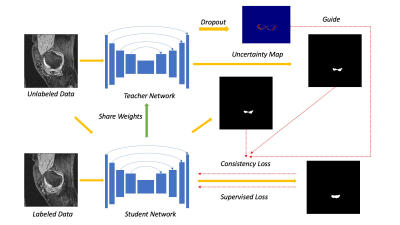
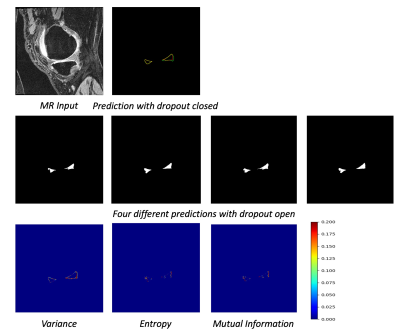
Yu et al. introduced an uncertainty-aware model by exploiting the uncertainty information for left atrium segmentation. 1 In Yu’s approach, the student model gradually learns from reliable pseudo predictions from the teacher model supervised by a segmentation loss and a consistency loss. The consistency loss is applied for comparing the predictions from the student and the teacher network on unlabeled data. With the guidance of uncertainty estimation, the consistency loss improves the teacher network with more reliable predictions. 1 In order to estimate uncertainty, adding Monte Carlo (MC) dropout to neural networks is an essential way for Bayesian networks approximation. 3 The sampling times T forward passes through the neural network during training to get multiple predictions. In this work, we evaluate this semi-supervised model in meniscus segmentation. The proposed model is tested on OAI Imorphics dataset which is a knee 3D-DESS MRI dataset including 88 subjects with the manual segmentation of meniscus. Datasets are randomly split subject-wise into the train and test subsets.Compared with Yu's work 1, U-Net 4 is applied as our student and teacher network backbone instead. We take Bayesian Segnet 5 as a reference to adopt dropout layers into U-Net with dropout rate 0.5. Rather than input both labeled and unlabeled data into the teacher network, we just feed unlabeled data into the teacher network. This is to ensure the uncertainty estimate feedback is totally from unannotated data. Our framework for meniscus segmentation can be illustrated in Figure 1. The weighting factor for balancing supervised loss and consistency loss is replaced by function $$$\alpha(E)=0.25*e^{(-5(1-E/E_{max}))^2}$$$. E denotes the current epoch and $$$E_{max}$$$ is the maximum epoch. 1 We take T as 10 to estimate the uncertainty and set the uncertainty threshold a fixed value 0.8. The stochastic gradient descent optimizer with weight decay 0.0001 was used during the training process. The models were built using the open-source PyTorch platform and were running on two Titan V GPU cards. To investigate the effect of different uncertainty measures on network performance, we experimented three different approaches to approximate uncertainty, including predicted sample variance, predictive entropy, and predictive mutual information. 2
Results
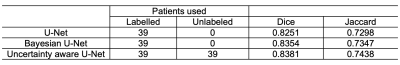
From OAI Imorphics dataset, we used 39 patients as the labeled data and another 39 patients as the unlabeled data for training. The remaining 10 patients are used as the test subset. Dropout was used during the test phase and sample T was 4 when calculating uncertainties. Figure 2 shows an example of uncertainties calculated using three approaches of sample variance, entropy, and mutual information, respectively. The Dice coefficients of networks with sample variance and predictive entropy are 0.8381 and 0.8317, respectively, while the uncertainty estimation of mutual information drives the Bayesian network to divergence. The high value on the uncertainty map with sample variance reflects the best unreliability prediction of this approach. Therefore, we choose sample variance as the uncertainty measure for our model in all our experiments.We used the Dice coefficient and Jaccard metric to quantitatively evaluate the performance of U-Net, Bayesian U-Net and our semi-supervised model. Figure 3 shows the segmentation performance of different methods. Both U-Net and Bayesian U-Net were trained on the same amount of labeled data. Compared with U-Net, the Bayesian U-Net with dropout layers for uncertainty estimation improves the segmentation performance from 0.8251 to 0.8354. The uncertainty-aware model proposed by us improves segmentation performance from 0.8251 to 0.8381 by the usage of unannotated data.
Discussion and Conclusions
In this work, we investigated the dropout-based Bayesian semi-supervised network for meniscus segmentation using MRI images. Our preliminary results indicates that the inclusion of the unannotated data with uncertainty estimation has the potential to improve the meniscus segmentation.Acknowledgements
This study is supported by a grant from the Innovation and Technology Commission of the Hong Kong SAR (Project MRP/001/18X) and a grant from the Research Grants Council of the Hong Kong SAR (Project SEG CUHK02).References
[1] Yu, L., Wang, S., Li, X., Fu, C. W., & Heng, P. A. (2019, October). Uncertainty-aware self-ensembling model for semi-supervised 3D left atrium segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention(pp. 605-613). Springer, Cham.
[2] Nair, T., Precup, D., Arnold, D. L., & Arbel, T. (2020). Exploring uncertainty measures in deep networks for multiple sclerosis lesion detection and segmentation. Medical image analysis, 59, 101557.
[3] Kendall, A., & Gal, Y. (2017). What uncertainties do we need in bayesian deep learning for computer vision?. In Advances in neural information processing systems (pp. 5574-5584).
[4] Ronneberger, O., Fischer, P., & Brox, T. (2015, October). U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical image computing and computer-assisted intervention (pp. 234-241). Springer, Cham.
[5] Kendall, A., Badrinarayanan, V., & Cipolla, R. (2015). Bayesian segnet: Model uncertainty in deep convolutional encoder-decoder architectures for scene understanding. arXiv preprint arXiv:1511.02680.
Figures