0796
QSMART: a parallel stage QSM pipeline for suppression of cortical and venous artifacts1Melbourne Brain Centre Imaging Unit, The University of Melbourne, Melbourne, Australia, 2Department of Biomedical Engineering, The University of Melbourne, Melbourne, Australia, 3Melbourne Neuropsychiatry Centre, The University of Melbourne, Melbourne, Australia, 4Department of Medicine and Radiology, The University of Melbourne, Melbourne, Australia, 5Department of Radiology, Royal Melbourne Hospital, Melbourne, Australia, 6Department of Neuroscience, Central Clinical School, Monash University, Melbourne, Australia, 7Department of Radiology, Alfred Hospital, Melbourne, Australia
Synopsis
We propose a two-stage QSM method, Quantitative Susceptibility Mapping Artifact Reduction Technique (QSMART), in which tissue and vein susceptibility values are estimated in parallel by generating a vasculature mask from the magnitude data using a Frangi filter. Spatially Dependent Filtering is employed for the background field removal and the two susceptibility estimates are combined in the final QSM map. QSMART is compared to RESHARP/iLSQR and V-SHARP/iLSQR inversion on 7T in vivo single and multiple-orientation scans. QSMART demonstrates superior artifact suppression in the cortex and near vasculature, and is a robust tool for susceptibility estimation. QSMART code is available at https://github.com/MBCIU/QSMART.
Introduction
Quantitative susceptibility mapping (QSM) is a novel MR technique that allows the mapping of tissue susceptibility values from MR phase images. QSM is an ill-conditioned inverse problem and in the presence of a wide range of susceptibility sources, streaking artifacts appear around high susceptibility regions, contaminating the QSM map. Serial two-stage inversion QSM methods1,2 identify high-susceptibility regions by thresholding a first-stage QSM map, and use the second stage to estimate low susceptibility sources. In this work, we propose a QSM pipeline consisting of two parallel stages of spatially dependent filtering (SDF) followed by iLSQR3. In QSMART, the susceptibility of the tissue and high susceptibility sources, identified by application of a Frangi filter, are estimated separately and combined to form the final QSM map.Methods
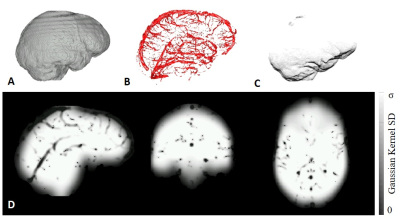
QSMART utilizes 3D generalized spatially dependent filtering (SDF)4 to remove the background field effects from the field shifts, by performing Gaussian filtering with spatially varying parameters. SDF suppresses cortical surface artifacts and the streaking artifacts around veins. The 3D SDF employs three masks: a brain mask, $$$M_{brain}$$$, generated using FSL's BET5; a vasculature mask derived by applying a Frangi filter6 on magnitude images, $$$M_{vein}$$$; and an indentation mask, $$$M_{Indent}$$$ to reduce artifacts near indented cortical surfaces such as the medial orbital gyrus. The indentation mask is generated by calculating the Gaussian curvature of the cortical surface (Figure 1A-C). Three proximity maps are defined based on the masks:1) a cortical proximity map, $$P_C=(F_{\sigma_C}*M_{Brain})M_{Brain},$$ where $$$F$$$ is a 3D Gaussian filter with standard deviation, $$$\sigma_C$$$.
2) a venous proximity map, $$P_V=\left(F_{\sigma_V}*(1-M_{Vein})\right)(1-M_{Vein}),$$
3) an indentation map,$$P_I=C_sF_{\sigma_I}*(1-M_{Indent}),$$ where $$$C_s$$$ is a scaling constant. The combined proximity map is a voxelwise multiplication of the three maps: $$P=P_{B}P_{V}P_{I}.$$ A map of spatially dependent standard deviations is given by $$$\alpha=\sigma \times (P^n)_2 M_{Brain}$$$, where the element-wise exponentiation, $$$(P^n)_2$$$, is rounded to 2 decimal places to accelerate the adaptive filtering operation. $$$~n~$$$is chosen such that $$$\alpha=1$$$ when $$$P=0.5$$$, giving an ordered set, $$$A = \{\alpha_1,...,\alpha_{101}\}$$$ consisting of all possible rounded values of $$$\alpha$$$. An index map, $$$X$$$, describes the spatial distribution of $$$\alpha$$$ values (Figure 1D). A set of Gaussian filtered images, $$$\phi$$$, is constructed from $$$A$$$. The background field is estimated by indexing $$$\phi$$$ using $$$X$$$.
A first stage susceptibility estimate is generated for all the voxels in the brain mask using iLSQR. In parallel, a second stage susceptibility map is constructed by removing the vasculature from the brain mask. The two susceptibility estimates are optimally combined2 to yield the QSMART map.
Experimental data: A healthy subject was scanned on a 7T Siemens MAGNETOM scanner (Erlangen, Germany) with a 1Tx/32Rx head coil (Nova Medical Inc.) using a 3D unipolar multi-echo Gradient-Recalled-Echo: TE = 4.80, 8.35, 11.90, 15.45 ms, TR = 18 ms, BW = 682 Hz/pix, FA = 9$$$^{\circ}$$$, resolution = 0.6$$$\times$$$0.6$$$\times$$$0.6 mm, matrix size = 334$$$\times$$$386$$$\times$$$224, GRAPPA = 2. The subject was scanned once in 5 different head orientations for COSMOS reconstruction. The scanning duration for each acquisition was 8.50 minutes.
The phase images of the coils were combined7 and unwrapped using a Laplacian-based phase unwrapping technique. The total field shift was estimated by fitting the echoes using a magnitude-weighted Least Squares method and normalized by the main magnetic field.
Results
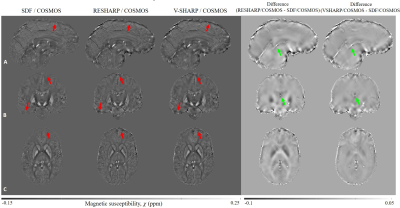
Figure 2 shows the single orientation results generated by QSMART, RESHARP/iLSQR, and V-SHARP/iLSQR. Cortical surface artifacts (red arrows) and x-shaped artifacts caused by veins (green arrows) are successfully suppressed by QSMART.Figure 3 shows the multiple orientation results generated by SDF/COSMOS, RESHARP/COSMOS, and V-SHARP/COSMOS. Cortical artifacts are significantly reduced by SDF/COSMOS (red arrows). Although COSMOS inversion reduces streaking artifacts by using multiple orientation data, the contribution of SDF in suppressing streaking artifacts near veins is shown in the difference images (green arrows).
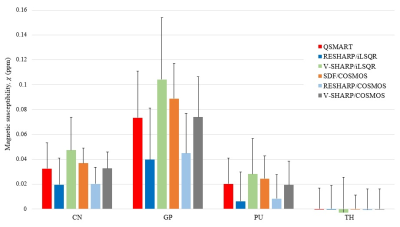
A region-of-interest analysis of susceptibility estimates in Caudate Nucleus (CN), Globus Pallidus (GP), Putamen (PU), and Thalamus (TH) is shown in Figure 4. QSMART and SDF/COSMOS show smaller variances compared to other single orientation and multiple orientation methods, respectively. The results of QSMART are similar to V-SHARP/COSMOS (p-value$$$>0.05$$$), demonstrating QSMART's ability as a single orientation pipeline to achieve a susceptibility distribution similar to a multiple orientation pipeline in the subcortical grey matter.
Discussion
QSMART implements two independent inversions in parallel by identifying vasculature from magnitude images using a Frangi filter, eliminating the need to use the first susceptibility map to determine high susceptibility veins1,2. The vasculature mask in QSMART can be adapted to incorporate pathological structures, such as ischaemic haemorrhage, lesions or haematoma, by changing the parameters in the Frangi filter accordingly. However, segmenting larger pathological structures may result in reduced coverage in the second QSM inversion 8, which may introduce inaccuracies in susceptibility estimation. QSMART has been validated on 3T and 7T human data, 4.7T mouse and 9.4T rat data (data not shown) with robustness of the artifact suppression across field strengths and brain types.Conclusion
QSMART produces susceptibility maps that robustly outperform similar methods, through suppression of streaking artifacts around veins and suppression of artifact at the cortical surface. Consistent generation of clean QSM maps such as those output by QSMART is vital in the clinical translation of QSM.Acknowledgements
We acknowledge the facilities, the scientific and technical assistance of the Australian National Imaging Facility, a National Collaborative Research Infrastructure Strategy (NCRIS) capability, at the Melbourne Brain Centre Imaging Unit of the University of Melbourne. The work was also supported by a research collaboration agreement with Siemens Healthineers.References
1. Wei, Hongjiang, et al. "Streaking artifact reduction for quantitative susceptibility mapping of sources with large dynamic range." NMR in Biomedicine 28.10 (2015): 1294-1303.
2. Sun, Hongfu, et al. "Quantitative susceptibility mapping using a superposed dipole inversion method: application to intracranial hemorrhage." Magnetic Resonance in Medicine 76.3 (2016): 781-791.
3. Li, Wei, et al. "A method for estimating and removing streaking artifacts in quantitative susceptibility mapping." Neuroimage 108 (2015): 111-122.
4. Ng, Amanda, et al. "Spatially dependent filtering for removing phase distortions at the cortical surface." Magnetic Resonance in Medicine 66.3 (2011): 784-793.
5. Smith, Stephen M. "Fast robust automated brain extraction." Human Brain Mapping 17.3 (2002): 143-155.
6. Frangi, Alejandro F., et al. "Multiscale vessel enhancement filtering." International conference on medical image computing and computer-assisted intervention. Springer, Berlin, Heidelberg, 1998.
7. Sun, Hongfu, et al. "Extracting more for less: multi‐echo MP2RAGE for simultaneous T1‐weighted imaging, T1 mapping, mapping, SWI, and QSM from a single acquisition." Magnetic Resonance in Medicine 83.4 (2020): 1178-1191.
8. Karsa, Anita, et al. "The effect of low resolution and coverage on the accuracy of susceptibility mapping." Magnetic Resonance in Medicine 81.3 (2019): 1833-1848.
Figures