0734
A Magnetic Resonance Imaging Simulation Framework of the Developing Fetal Brain1Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 2CIBM Center for Biomedical Imaging, Lausanne, Switzerland, 3Advanced Clinical Imaging Technology (ACIT), Siemens Healthcare, Lausanne, Switzerland, 4Signal Processing Laboratory 5 (LTS5), Ecole Polytechnique Fédérale de Lausanne (EPFL), Lausanne, Switzerland
Synopsis
Accurate characterization of in utero human brain maturation is critical. However, the limited number of exploitable magnetic resonance acquisitions not corrupted by motion in this cohort of sensitive subjects hinders the validation of advanced image processing techniques. Numerical simulations can mitigate these limitations by providing a controlled environment with a known ground truth. We present a flexible framework that simulates magnetic resonance acquisitions of the fetal brain in a realistic setup including stochastic motion. From simulated images comparable to clinical acquisitions, we assess the accuracy and robustness of super-resolution fetal brain magnetic resonance imaging with respect to noise and motion.
Introduction
There is a growing awareness of the importance of processes occurring during early brain development on health later in life 1,2. Magnetic Resonance Imaging (MRI) is a powerful tool for depicting brain tissue contrast and therefore has the potential to investigate equivocal neurological patterns in utero. Before birth, fast 2D spin echo sequences are typically used to minimize the effects of unpredictable fetal motion during the acquisition. Access to large-scale data, usually corrupted by stochastic movements of the fetus, remains relatively scarce, hampering the development and evaluation of advanced image processing methods. Numerical phantoms provide a controlled environment where the ground truth is known to meet a variety of challenges related to the acquisition process for accurate, robust and reproducible research 3,4. In this context, we have developed a simulation framework for Half-Fourier Acquisition Single-shot Turbo spin Echo (HASTE). Since motion correction is crucial in fetal MRI, the potential of this new tool is highlighted by an application example: the assessment of super-resolution (SR) reconstruction techniques that combine orthogonal series of 2D thick slices into an isotropic 3D high-resolution volume of the fetal brain with reduced intensity artefacts and motion sensitivity 5–9.Methods
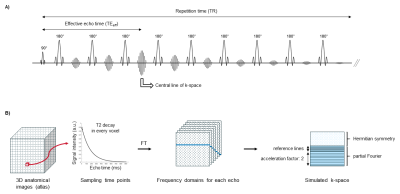
Figure 1 provides an overview of the simulation pipeline of HASTE acquisitions implemented using MATLAB (MathWorks, R2019a). Segmented high-resolution anatomical images from a normative spatiotemporal MRI atlas 10 are used as a model of the normal fetal brain. Segmented brain tissues are labeled as gray matter, white matter or cerebrospinal fluid, and are assigned corresponding T1 and T2 relaxation times at 1.5T 11–15. The extended phase graph (EPG) simulation 16 allows for computing the decay of the transverse magnetization over time in every voxel of the anatomical images from the reference T1 and T2 maps and from simulated intensity non-uniformities 17, according to the HASTE sequence pulse design. The Fourier transform of the resulting 4D matrix is used to sample the actual k-space of the simulated HASTE images as described in Figure 2. While intra-slice motion is neglected, inter-slice random rigid movements of the fetus are implemented during k-space sampling according to motion estimation from clinical data 18. Three levels of motion are defined: low, moderate and strong, that are characterized by less than 5%, 10% and 20% of corrupted slices respectively, and an amplitude of translation in every direction of [-1, 1]mm, [-2, 2]mm and [-2, 2]mm and of rotation of [-2, 2]°, [-4, 4]° and [-4, 4]° respectively. Complex Gaussian noise is added to simulate thermal noise during the acquisition. The final simulated HASTE images are reconstructed by 2D inverse Fourier transform. HASTE acquisitions of the fetal brain are simulated in three orthogonal orientations with a shift of ± 1.6 mm in-plane for acquisitions in the same orientation. The acquisition parameters comply with the clinical protocol set up at our local hospital for fetal brain examination. A case study on SR is presented using a previously reported reconstruction pipeline 8,19. The quality of SR reconstruction is evaluated based on the normalized root mean squared error (NRMSE), the local structural similarity (SSIM) index 20 and its mean (MSSIM) over the image compared to a 3D 1.1-mm isotropic HASTE simulation uncorrupted by noise or movement. Six realizations are simulated for each SNR. The impact of low, respectively moderate movements of the fetus on the quality of SR reconstruction is studied using a reference series without motion, respectively with low motion amplitude.Results
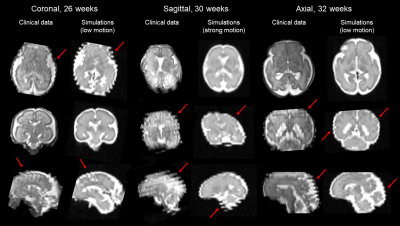
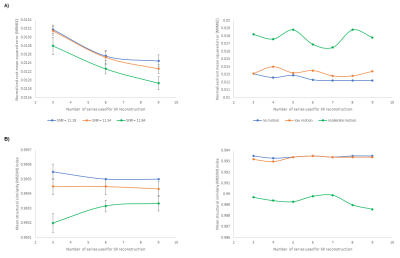
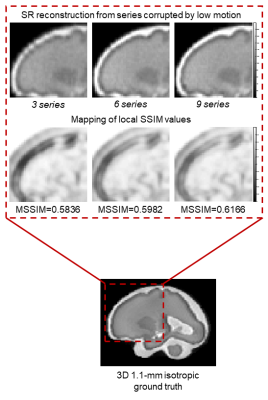
Figure 3 compares simulated motion-corrupted HASTE images of the fetal brain at 26, 30 and 32 weeks of gestational age to clinical acquisitions. Figure 4 shows the NRMSE and the MSSIM between SR reconstructions from various numbers of low-resolution series and a 3D isotropic high-resolution reference. The NRMSE decreases when increasing the number of series used for SR reconstruction. Noisier images lead to a slight decrease in the MSSIM, which in turn increases with the number of series. The quality of SR reconstructions from simulated data with an SNR similar to that observed in clinical acquisitions is the same as for SR reconstructions from simulated data with twice as much signal. The addition of motion-corrupted low-resolution series to reconstruct a SR volume of the fetal brain does not increase the MSSIM. In the case of moderate motion, the MSSIM is smaller than in the case of low motion. Figure 5 illustrates the improved sharpness in a region-of-interest of SR reconstruction when increasing the number of motion-corrupted series at an SNR similar to that of clinical acquisitions.Discussion and Conclusion
We developed a new framework for fetal brain MRI simulations and explored its potential for evaluation and optimization of SR reconstruction techniques. This powerful tool simulates HASTE images comparable to real clinical acquisitions. Its flexibility in the choice of the sequence parameters but also other settings such as the gestational age makes it possible to simulate MR images of the fetal brain throughout the growth of the fetus with various SNR and amplitude of fetal movements. The controlled environment implemented demonstrates that the SR reconstruction algorithm used is robust to noise and motion. Such a numerical phantom provides a valuable framework for reproducibility studies and validation of advanced image processing techniques.Acknowledgements
This work was supported by the Swiss National Science Foundation through grants 141283 and 182602, the Centre d’Imagerie BioMédicale (CIBM) of the UNIL, UNIGE, HUG, CHUV and EPFL, and the Leenaards and Jeantet Foundations.References
1. Kwon EJ, Kim YJ. What is fetal programming?: a lifetime health is under the control of in utero health. Obstet Gynecol Sci. 2017;60(6):506-519. doi:10/gf5fj6
2. O’Donnell KJ, Meaney MJ. Fetal origins of mental health: The developmental origins of health and disease hypothesis. Am J Psychiatry. 2017;174(4):319-328. doi:10/f92t6n
3. Wissmann L, Santelli C, Segars WP, Kozerke S. MRXCAT: Realistic numerical phantoms for cardiovascular magnetic resonance. J Cardiovasc Magn Reson. 2014;16:63. doi:10/gf6bpx
4. Roy CW, Marini D, Segars WP, Seed M, Macgowan CK. Fetal XCMR: a numerical phantom for fetal cardiovascular magnetic resonance imaging. Journal of Cardiovascular Magnetic Resonance. 2019;21(1):29. doi:10/gf33j3
5. Rousseau F, Kim K, Studholme C, Koob M, Dietemann JL. On super-resolution for fetal brain MRI. International Conference on Medical Image Computing and Computer-Assisted Intervention. 2010;13(Pt 2):355-362. doi:10/bns47p
6. Gholipour A, Estroff JA, Warfield SK. Robust super-resolution volume reconstruction from slice acquisitions: application to fetal brain MRI. IEEE Transactions on Medical Imaging. 2010;29(10):1739-1758. doi:10/b2xmdp
7. Kuklisova-Murgasova M, Quaghebeur G, Rutherford MA, Hajnal JV, Schnabel JA. Reconstruction of fetal brain MRI with intensity matching and complete outlier removal. Medical Image Analysis. 2012;16(8):1550-1564. doi:10/f4hxwz
8. Tourbier S, Bresson X, Hagmann P, Thiran J-P, Meuli R, Cuadra MB. An efficient total variation algorithm for super-resolution in fetal brain MRI with adaptive regularization. NeuroImage. 2015;118:584-597. doi:10/f7p5zx
9. Kainz B, Steinberger M, Wein W, et al. Fast volume reconstruction from motion corrupted stacks of 2D slices. IEEE Transactions on Medical Imaging. 2015;34(9):1901-1913. doi:10/f3svr5
10. Gholipour A, Rollins CK, Velasco-Annis C, et al. A normative spatiotemporal MRI atlas of the fetal brain for automatic segmentation and analysis of early brain growth. Sci Rep. 2017;7(1):476. doi:10/gf39sn
11. Hagmann CF, De Vita E, Bainbridge A, et al. T2 at MR imaging is an objective quantitative measure of cerebral white matter signal intensity abnormality in preterm infants at term-equivalent age. Radiology. 2009;252(1):209-217. doi:10/bqkd9r
12. Blazejewska AI, Seshamani S, McKown SK, et al. 3D in utero quantification of T2* relaxation times in human fetal brain tissues for age optimized structural and functional MRI. Magn Reson Med. 2017;78(3):909-916. doi:10/gf2n9z
13. Vasylechko S, Malamateniou C, Nunes RG, et al. T2* relaxometry of fetal brain at 1.5 Tesla using a motion tolerant method. Magnetic Resonance in Medicine. 2015;73(5):1795-1802. doi:10/gf2pbh
14. Nossin-Manor R, Card D, Morris D, et al. Quantitative MRI in the very preterm brain: Assessing tissue organization and myelination using magnetization transfer, diffusion tensor and T1 imaging. Neuroimage. 2013;64:505-516. doi:10/f4jgtg
15. Yarnykh VL, Prihod’ko IY, Savelov AA, Korostyshevskaya AM. Quantitative assessment of normal fetal brain myelination using fast macromolecular proton fraction mapping. American Journal of Neuroradiology. 2018;39(7):1341-1348. doi:10/gdv9nf
16. Weigel M. Extended phase graphs: Dephasing, RF pulses, and echoes - pure and simple. Journal of Magnetic Resonance Imaging. 2015;41(2):266-295. doi:10/gghctd
17. BrainWeb: Simulated brain database. https://brainweb.bic.mni.mcgill.ca/brainweb/
18. Oubel E, Koob M, Studholme C, Dietemann J-L, Rousseau F. Reconstruction of scattered data in fetal diffusion MRI. Medical image analysis. 2012;16(1):28-37. doi:10/fhbqtj
19. Tourbier S, Bresson X, Hagmann P, Meuli R, Bach Cuadra M. Sebastientourbier/Mialsuperresolutiontoolkit: MIAL Super-Resolution Toolkit v1.0. Zenodo; 2019. doi:10.5281/zenodo.2598448
20. Wang Z, Bovik A, Sheikh H, Simoncelli E. Image quality assessment: From error visibility to structural similarity. IEEE Transactions on Image Processing. 2004;13:600-612. doi:10/c7sr27
Figures