0729
Open-Source MR Imaging Workflow1MR Physics, German Center for Neurodegenerative Diseases (DZNE), Bonn, Germany, 2MR R&D Collaborations, Siemens Medical Solutions USA Inc., Chicago, IL, United States, 3Department of Radiology - Medical Physics, University Medical Center Freiburg, Freiburg, Germany, 4Department of Physics & Astronomy, University of Bonn, Bonn, Germany
Synopsis
We demonstrate an open-source MR imaging workflow for MR sequence development and online image reconstruction. The open-source approach improves vendor-independent reproducibility of new or modified MR sequences and reconstruction algorithms. An extensible online reconstruction pipeline allows for fast evaluation of the acquired data on the scanner directly. The pipeline supports implementation of advanced open-source image reconstruction techniques and is not restricted to a specific sequence. We successfully implemented an open-source spiral sequence with Pulseq for execution at a 7T MR scanner. A non-Cartesian image reconstruction was done with the BART toolbox using GIRF trajectory prediction.
Introduction
In recent years, open-source pulse sequence programming has grown with the development of frameworks like Pulseq1 and TOPPE2. These frameworks allow for rapid prototyping and better reproducibility of MR sequences on any platform through open-source file formats and low level representations of the pulse sequence. For the design of pulse sequences, high-level scripting languages such as Python or Matlab, or graphical user interfaces as for example JEMRIS3, are available.On the other end of the imaging process, several open-source tools exist for image reconstruction4. However, these tools are often used offline as it is complicated to integrate them into the scanners reconstruction pipeline.
In our work, we demonstrate an end-to-end open source imaging workflow from acquisition to reconstruction, using a vendor provided implementation of the ISMRM raw data (ISMRMRD) format5 for inline integration. As an example, data is acquired with a spiral sequence and reconstructed online at the MR scanner.
Methods
The following workflow (Figure 1) was tested with an interleaved spiral-out gradient echo acquisition at a prototype MAGNETOM 7T scanner (Siemens Healthcare).MR sequences are designed in Python, mainly with the PyPulseq6 package, which readily provides support for standard RF pulses and gradient waveforms. For the readout gradients, a time-optimized Archimedean spiral is used, calculated with code by Lustig et. al.7. All sequence blocks are exported to the Pulseq file, containing the complete sequence definition.
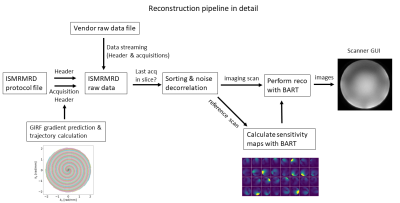
As Pulseq offers only a limited possibility to save sequence-specific protocol information in the sequence file, an addiditional protocol file in the ISMRMRD format is created. This protocol contains all header and acquisition information necessary for the reconstruction. This includes k-space counters, flags and the nominal readout gradients. Both sequence and protocol files are exported to the scanner. The sequence is selected in the scanner GUI and executed by a vendor-specific interpreter, which supports integrated field-of-view positioning.
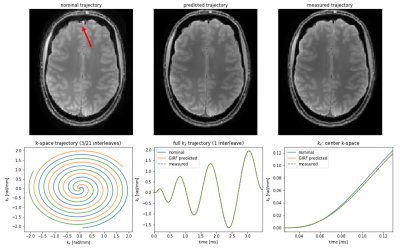
Online image reconstruction is performed in real-time with the acquisition using the prototype Framework for Image Reconstruction Environments (FIRE) interface, the Siemens implementation of the streaming ISMRMRD format. In the streaming process, the data is converted to the ISMRMRD format and transfered to a Python reconstruction server. In the first step of the reconstruction pipeline (Figure 2), the protocol information is mirrored from the protocol file to the acquisition file. This is accompanied by the prediction of the actual gradient waveform from the nominal gradient with the Gradient Impulse Response Function (GIRF)8, which is well-suited for spiral imaging at 7T9. The resulting k-space trajectory is added to the raw data. For comparison, the actual k-space trajectory was measured using a field-camera (Skope Magnetic Resonance Technologies)10.
Acquisitions are collected from the streamed data until k-space data for one slice is acquired completely. Incoming data is sorted and optional noise prewhitening is performed. Image reconstruction is performed with a non-Cartesian parallel imaging reconstruction from the Berkeley Advanced Reconstruction Toolbox (BART)11. For accelerated acquisitions, sensitivity maps are calculated with BART from a Cartesian reference scan preceding the spiral scan.
Reconstructed images are sent back to the scanner GUI for visualisation. Optionally, the reconstruction pipeline can also be executed offline, independently from the vendors' interface.
Results
Reconstructed images are shown in Figure 3. Reconstructions with GIRF-predicted trajectories are compared to reconstructions with nominal and measured trajectories. The GIRF-predicted trajectory closely follows the measured (bottom row) leading to comparable reconstructed images. The reconstructed image using the nominal trajectory is slightly rotated and shows minor artifacts.Discussion
The pulse sequence design with Pulseq in Python is straightforward and may require less effort compared to the vendors' sequence programming environment. It allows for the use of multiple open-source tools for the design of gradients and RF pulses. However, special attention must be given to the hardware-specific sequence requirements, as RF, gradient and ADC limits must be set manually in Pulseq.The online reconstruction pipeline enables a fast visual evaluation of the acquired spiral data and is extensible by adding additional reconstruction or post-processing algorithms and other open-source tools. Once the streaming ISMRMRD interface is set up, advanced image reconstruction techniques are easily implemented. Reconstruction of the spiral data with the BART toolbox resulted in high-quality images, which was further improved by GIRF-based trajectory prediction. The reconstruction is not restricted to spiral trajectories though, as the ISMRMRD format is customizable with any user-defined trajectories and parameters. Furthermore, it is possible to integrate image correction or analysis tools into the pipeline.
Pulseq sequence files and protocol files can be published along with imaging results, which is expected to improve multi-site reproducibility on scanners from different vendors or from the same vendor running different software versions. Reproducibility can be improved even further by sharing the image reconstruction pipeline.
Conclusion
The demonstrated end-to-end open-source sequence programming and image reconstruction workflow allows for rapid prototyping and testing of new or modified MR sequences. The online image reconstruction pipeline is versatile as it is not restricted to any type of MR sequence and can be extended in various ways by own code or other open source tools. Sequence development and image reconstruction pipelines can be shared freely as they are vendor independent, which can significantly improve the reproducibility of MR experiments.Acknowledgements
No acknowledgement found.References
1. Layton, K. J. et. al. Pulseq: A rapid and hardware‐independent pulse sequence prototyping framework, MRM, 2017;77(4):1544-1552
2. Nielsen, J. F. & Noll, D. C. TOPPE: A framework for rapid prototyping of MR pulse sequences, MRM, 2018;79(6):3128-3134
3. Stöcker, T. et. al. High-performance computing MRI simulations. MRM, 2010;64(1):186–93
4. An overview of available tools can be found on https:https://www.opensourceimaging.org/projects.
5. Inati, J. I. et. al. ISMRM Raw data format: A proposed standard for MRI raw datasets, MRM, 2017;77(1):411-421
6. Ravi, K. et. al PyPulseq: A Python Package for MRI Pulse Sequence Design. J. Open Source Softw. 2019;4(42):1725
7. Lustig, M. A fast method for designing time-optimal gradient waveforms for arbitrary k-space trajectories. IEEE Trans Med Imaging. 2008;27(6):866-73.
8. Vannesjo, S. J. et al. Gradient System Characterization by Impulse Response Measurements with a Dynamic Field Camera. MRM 2013;69:583-593
9. Veldmann, M. at. al. Spiral in/out trajectory with GIRF prediction for passband bSSFP fMRI at 7T, ESMRMB 2020 Virtual Meeting
10. Barmet, C. et. al. Spatiotemporal Magnetic Field Monitoring for MR, MRM 2008;60:187–197
11. Uecker, M. et. al. Berkeley Advanced Reconstruction Toolbox. Annual Meeting ISMRM, Toronto, Proc. Intl. Soc. Mag. Reson. Med. 2015;23:2486, DOI: 10.5281/zenodo.592960
Figures