0728
TIRL: Automating Deformable Slice-to-Volume Registration Between Stand-Alone Histology Sections and Post-Mortem MRI
Istvan N Huszar1, Karla L Miller1, and Mark Jenkinson1
1Wellcome Centre for Integrative Neuroimaging, FMRIB, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom
1Wellcome Centre for Integrative Neuroimaging, FMRIB, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, United Kingdom
Synopsis
An open-source framework (TIRL) and a novel 3-stage pipeline are presented to automate the registration between sparsely sampled small histology sections and 3-D MRI data for validating imaging biomarkers. In addition to affine registration, deformable slice-to-volume registration is employed to compensate for both in-plane and through-plane distortions of the histology sections. Each stage of the pipeline is shown to achieve submillimetre precise alignments, surpassing the accuracy of previous methods. With photographic intermediaries the pipeline is fully automatic and does not depend on serial histological sectioning or specialist cutting and stain automation hardware. The tools are provided as part of FSL.
Introduction
There is an increasing demand for non-invasive biomarkers that can reliably indicate the presence and extent of neurodegeneration in its early stages. Various MRI methods have shown great potential to indicate the event of upper motor neuron involvement in the progression of motor neuron disease (MND) at the population level, but these have not been systematically validated against traditional histological markers at the individual level1. Validation relies on a precise and preferably automatic method to align a large number of MRI and histology images of the same tissue, which poses unique challenges compared to more conventional MRI-to-MRI registration. Here we present a novel image registration framework (Tensor Image Registration Library, TIRL) and methodological validation of two registration pipelines for aligning conventional sparsely sampled histology sections with post-mortem 3-D MRI data through photographic intermediaries.Materials and Methods
TIRL was designed and implemented to be compatible with a wide range of images (including histology, MRI, and photographic formats, 2-D and 3-D images, single- and multi-channel images), has native support for slice-to-volume registration, and allows fast prototyping of bespoke registration pipelines. Details of the implementation have been summarised elsewhere2 and the framework was released with FSL 6.0.4.3Here we performed experiments to validate the accuracy of two TIRL-based registration pipelines on a multi-modal imaging dataset that was collected from 16 post-mortem human brains as part of a larger MND study4 (Figure 1). The formalin-fixed brains were scanned using a steady-state free-precession sequence (TRUFI) at 0.25 mm/voxel isotropic resolution at 7T, and dissected by hand to isolate approximately 25x30 mm large tissue blocks for multi-modal histology. Motor blocks were excised first. The brain was subsequently sliced into 1-cm thick coronal sections and the extramotor blocks were extracted from these. The tissue blocks and the coronal slices were photographed, yielding quasi-rigid intermediate reference images for the registration between histology and MRI: one (the block-face image) for the motor blocks and two (the block-face image and the brain slice photograph) for the extramotor blocks.
The 3-stage pipeline was used to register stained histology sections of the extramotor blocks to MRI space. Stage 1 employed a sequence of rotation search, rigid, and affine initialisation that was eventually refined by diffusion-regularised deformable registration minimising the MIND metric5. In Stage 2, the insertion sites for the tissue block were automatically inferred from cut-out whole-brain slice photographs (see Figure 1), and registered in place by the same sequence of operations as in Stage 1. Stage 3 refined initial estimates of the slice position and orientation, carried out 2D-to-3D affine registration, and used 32 quasi-random control points to optimise in-plane, then both in-plane and through-plane deformations of the slice to compensate for the imperfections of freehand slicing.
In the 2-stage pipeline, the slice-to-volume registration was carried out between the block-face image and the MRI volume, and the necessary initialisation was supplied manually (Figure 2).
The accuracy of the registration was thoroughly investigated in each case. In Stage 1, the grey-white matter boundaries (GWMB) were manually segmented on the original histology image and the block-face photo, and their mean distance (MCD) was measured after registration on 14 callosal and 14 hippocampal blocks. The results were compared against registrations by ANTs6 with Mattes and cross-correlation (CC) similarity metrics.
The accuracy of Stage 2 was evaluated on 87 blocks by visualising the alignment of perforating vessels and measuring MCDs. In Stage 3, the alignment was also visualised by GWMBs after each substage and estimated by registering synthetic slices that were extracted from the 3-D MRI data and pre-deformed by a quadratic polynomial. This experiment was repeated in the presence of Gaussian noise (SNR=10) to test for robustness.
Results
In Stage 1 (Figure 3), TIRL registered callosal samples with 0.25 mm MCD, and hippocampal sections with 0.4 mm MCD. For both types of blocks, higher registration errors were observed with ANTs with both the Mattes (0.42 mm, 0.7 mm) and the CC metrics (0.28 mm, 0.65 mm).In Stage 2 (Figure 3), the block matching process found the correct insertion site in 81/87 cases, and there were no visible errors in the registration. The registration error (MCD) was less than 0.2 mm in all cases, which was also supported by the close correspondence of perforating vascular structures.
In Stage 3 (Figure 4), significant through-plane deformations (0-6.5 mm) were necessary to compensate for the imperfections of freehand cutting. The distance of the synthetic slices from the registration target was 5-10 mm at the beginning, which fell below 0.2 mm by the end of the registration. Repeated experiments indicated robust convergence to the registration target and this behaviour was preserved in the presence of Gaussian noise as well.
The observed accuracies were preserved after combining stage-specific transformations to obtain end-to-end histology-to-MRI mapping (Figure 5) for all extramotor blocks. The alignment of the motor blocks by TIRL was more accurate than it was manually estimated by ROIs from the Juelich atlas (Figure 2).
Conclusion
The proposed pipelines provided robust and submillimetre accurate registration of sparsely sampled histology images to 3-D MRI data. The automation of this process allows MRI findings to be validated against histological ground truth in large disease cohorts.Acknowledgements
I.N.H. was supported by funding from the Engineering and Physical Sciences Research Council (EPSRC) and Medical Research Council (MRC) [grant number EP/L016052/1], the Clarendon Fund and the Chadwyck-Healey Charitable Trust at Kellogg College, Oxford, UK. The Wellcome Centre for Integrative Neuroimaging is supported by core funding from the Wellcome Trust (203139/Z/16/Z). The authors would like to further express their gratitude to the donors and benefactors of the Oxford Brain Bank.References
- Bede, P., The histological correlates of imaging metrics: postmortem validation of in vivo findings. Amyotrophic Lateral Sclerosis and Frontotemporal Degeneration, 2019. 20(7-8): p. 457-460.
- Huszar, I.N., et al., Tensor Image Registration Library: Automated Non-Linear Registration of Sparsely Sampled Histological Specimens to Post-Mortem MRI of the Whole Human Brain. bioRxiv, 2019: p. 849570.
- TIRL page of the FSLwiki: https://fsl.fmrib.ox.ac.uk/fsl/fslwiki/TIRL, accessed on 16 Dec 2020.
- Pallebage-Gamarallage, M., et al., Dissecting the pathobiology of altered MRI signal in amyotrophic lateral sclerosis: A post mortem whole brain sampling strategy for the integration of ultra-high-field MRI and quantitative neuropathology. BMC Neurosci, 2018. 19(1): p. 11.
- M.P., et al., MIND: modality independent neighbourhood descriptor for multi-modal deformable registration. Med Image Anal, 2012. 16(7): p. 1423-35. [6] Avants, B.B., et al., The Insight ToolKit image registration framework. Front Neuroinform, 2014. 8: p. 44.
- Avants, B.B., et al., The Insight ToolKit image registration framework. Front Neuroinform, 2014. 8: p. 44.
Figures
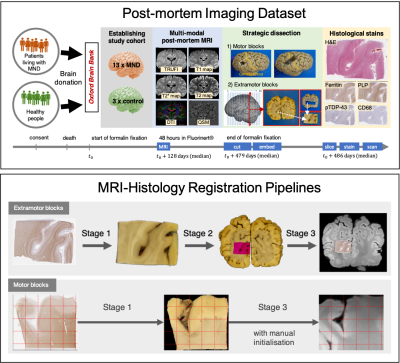
Figure 1. TOP: Acquisition
of post-mortem MRI, dissection photographs, and histology images. Motor and
extramotor blocks were sampled differently, yielding one or two photographic
intermediates (the block-face and the coronal slice images). BOTTOM: Histology-to-MRI
registration pipelines. Stage-specific transformations are combined to map the
high-resolution histology (0.5 mm/px) image into MRI space
(0.25 mm/vx). The MRI data is resampled by cubic spline interpolation to
produce a 2-D image.
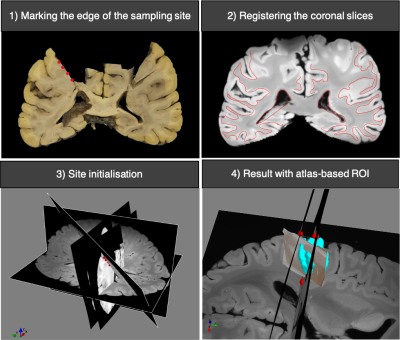
Figure 2. Histology-to-MRI
registration of a motor block using manual site initialisation. The edges of the sampling site are marked on two
consecutive coronal slices that are registered to MRI space by the stage-3
routine. The sampling site is initialised as the best-fit plane to the markers,
and the stage-3 routine is performed to find the exact sampling site. The
registered histology section is shown relative to a manually determined region
of interest (cyan) based on the FSL–Juelich atlas.
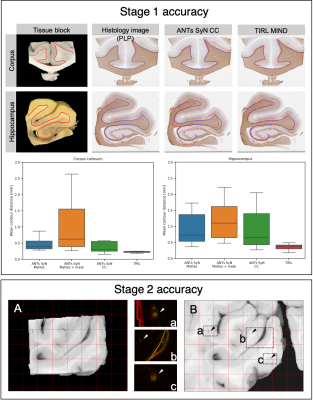
Figure 3. Accuracy 2D-to-2D
registration in Stages 2 and 3. STAGE 1: Representative
registration result showing the superior performance of TIRL versus ANTs
(Mattes and CC similarity metrics) in terms of the mean contour distance
between grey-white matter boundaries. STAGE 2: Highly accurate alignment
between the tissue block photo (A) and the coronal slice photo (B) showing
close correspondence of submillimetre-size perforating vessels. Grid spacing: 5
mm.
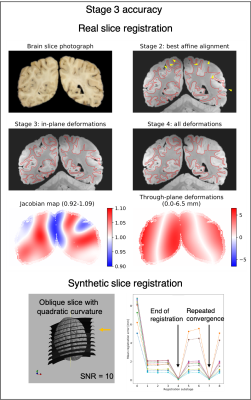
Figure 4. Accuracy of
slice-to-volume registration in Stage 3. TOP:
The alignment of the same coronal slice after successive substages of the
registration. The conservative Jacobian range suggests anatomically consistent
diffeomorphic transformation. The substantial through-plane deformations
effectively compensate for the imprecisions of free-hand slicing. BOTTOM:
Accurate registration of synthetic slices with repeated convergence to the
registration target. Each of the curves represents a single slice.
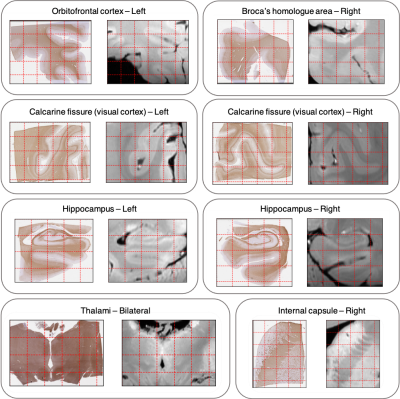
Figure 5. Representative side-by-side
comparison of histology and MRI data for a set of extramotor blocks. End-to-end registrations have uniform
submillimetre accuracy in all tested anatomical regions. Grid spacing: 5 mm (10
mm for the thalamus block).