0726
Physiopy: A community-driven suite of tools for physiological recordings in neuroimaging
Katherine Louise Bottenhorn1, Daniel Alcalà-Lopez2, Apoorva Ayyagari3, Molly G Bright4, César Caballero-Gaudes2, Inés Chavarria2, Vicente Ferrer2, Soichi Hayashi5, Vittorio Iacovella6, François Lespinasse7, Ross Davis Markello8, Stefano Moia2, Robert Oostenveld9,10, Taylor Salo1, Rachael Stickland4, Eneko Uruñuela2, Merel Margaretha van der Thiel11, and Kristina M Zvolanek12
1Department of Psychology, Florida International University, Miami, FL, United States, 2Basque Center on Cognition, Brain and Language, Donostia, Spain, 3Northwestern University, Chicago, IL, United States, 4Physical Therapy and Human Movement Sciences, Northwestern University, Chicago, IL, United States, 5Indiana University, Bloomington, IN, United States, 6CIMeC - Center for Mind / Brain Sciences, The University of Trento, Trento, Italy, 7Psychology, Université de Montréal, Montréal, QC, Canada, 8McGill University, Montréal, QC, Canada, 9Donders Institute for Brain, Cognition and Behaviour, Radboud University, Nijmegen, Netherlands, 10NatMEG, Karolinska Institutet, Stockholm, Sweden, 11Department of Radiology & Nuclear Medicine, Maastricht University Medical Center, Maastricht, Netherlands, 12Biomedical Engineering, Northwestern University, Chicago, IL, United States
1Department of Psychology, Florida International University, Miami, FL, United States, 2Basque Center on Cognition, Brain and Language, Donostia, Spain, 3Northwestern University, Chicago, IL, United States, 4Physical Therapy and Human Movement Sciences, Northwestern University, Chicago, IL, United States, 5Indiana University, Bloomington, IN, United States, 6CIMeC - Center for Mind / Brain Sciences, The University of Trento, Trento, Italy, 7Psychology, Université de Montréal, Montréal, QC, Canada, 8McGill University, Montréal, QC, Canada, 9Donders Institute for Brain, Cognition and Behaviour, Radboud University, Nijmegen, Netherlands, 10NatMEG, Karolinska Institutet, Stockholm, Sweden, 11Department of Radiology & Nuclear Medicine, Maastricht University Medical Center, Maastricht, Netherlands, 12Biomedical Engineering, Northwestern University, Chicago, IL, United States
Synopsis
Physiopy is a community-driven effort that aims to offer a full data preparation pipeline for non-neural physiological recordings in neuroimaging research and to build consensus on “best practices” among researchers of this specific domain. Its primary goal is to facilitate physiological data collection and sharing, according to an existing data standard and ontology. Corollary goals of the community are to provide recommendations and tools for (1) physiological data acquisition in the MR environment, (2) appropriate processing of these data, (3) organization of resulting datasets and meta-data, and (4) computing metrics for the removal of confounding physiological signals from fMRI data.
Introduction
The blood oxygenation level dependent (BOLD) functional magnetic resonance imaging (fMRI) signal inherently contains information from many physiological sources.1,2 This can confound our understanding of the neural activity captured by fMRI or provide complementary information concerning other biological processes. Recording peripheral physiological signals adds value to the neuroimaging data acquired, both for more stringent data cleaning, for studying physiologically-driven vascular effects, and for studying neural correlates of physiological variability. Physiological recordings are not systematically collected with fMRI data, in part due to a lack of established recommendations, protocols, and tools for collecting, processing, and sharing non-neural physiological recordings. Data sharing and harmonization across research groups are increasingly important, especially given recent work highlighting that the sample sizes required for well-powered brain-behavior analyses are infeasible for individual researchers to collect.3-5 Here, we introduce physiopy, a community-driven effort that aims to meet these needs by providing resources, recommendations, and software tools for collecting, organizing, and processing physiological recordings collected concurrently with fMRI data. Physiopy leverages the widely-used Brain Imaging Data Structure (BIDS)6 a community- and institution-endorsed (meta-)data organization scheme that is built into existing data sharing repositories, to facilitate and encourage data sharing. By providing this set of user-friendly tools, we aim to encourage researchers not only to include physiological measures in their studies, but also to share these data with the wider community; all in an open, transparent, and reproducible manner.Methods
Currently, Physiopy presents two software tools, as Python3 command-line interfaces, and documented recommendations for collecting physiological data.Physiopy’s first tool is phys2bids 7 which turns input physiological recordings (and optional meta-data) into BIDS-formatted output. Phys2bids currently accepts physiological recordings in three formats: AcqKnowledge native files (i.e., from a BIOPAC collection system), LabChart files (i.e., from ADInstruments collection), and MATLAB data files containing recordings from either of the two systems. Within these files, data are organized into “channels”, representing distinct physiological recordings and event signals such as photosensors and triggers. Before processing this input, a user can easily verify the contents of the file, retrieve channel names, sampling rates, and a figure plotting all the data.
Then, in processing the file, researchers can choose to simply transform the data from the entire neuroimaging session into separate compressed, tab-separated (i.e., .tsv.gz) files per sampling frequency (i.e. a BIDS compliant output with minimal processing). In minimal processing, phys2bids can also rely on a trigger channel, indicating duration of the (f)MRI acquisition(s) (or “runs”), and use this information to save recordings in run-specific output files (BIDS compliant as well). Specifics about the acquisition and recordings can be provided in other command-line arguments or in a separate heuristics file (also given as an argument), which will also be converted to a BIDS-formatted meta-data file, if provided. In this process, a report is generated, which includes interactive plots of each channel, the output directory structure and files, and logged pipeline output.
The second tool is phys2denoise, which provides physiological metrics and derived time series for removing non-neural physiological noise (i.e., “denoising) from simultaneously collected fMRI data. Phys2denoise accepts phys2bids-processed or physiological recording files, and computes cardiac and respiratory rates, respiratory pattern variability, and/or respiratory variance, per users’ specifications.
Physiopy documentation includes installation instructions for the aforementioned tools, creating the associated heuristics file(s), and recommendations for physiological data collection during fMRI studies. These recommendations include (1) motivations for including physiological recordings in a study, (2) a description of how cardiac pulsations, breathing, and blood gases are recorded, (3) the equipment recommended for acquiring these data, and (4) suggestions for the use of physiological recordings, once collected.
Results
Figure 2 shows the example output from phys2bids without any processing (i.e., using the “-info” option to inspect your data): 2A shows the logged channel names and sampling frequencies, while 2B shows the plotted data. Figure 3 shows the trigger estimation used in the phys2bids minimal processing. Figure 4 presents the interactive reporting output, displaying downsampled data from each channel for quality assurance purposes (4A), a print-out of the output directory containing processed files (4B), and phys2bids logging for clarity on the inner workings of phys2bids (4C).Conclusion
The goal of physiopy is threefold, developing a set of tools that encourage researchers to (1) appropriately collect and (2) share physiological recordings with their neuroimaging data, as well as (3) to provide a community dedicated to improving these tools and to reach consensus on best practices in neuroimaging research. Physiopy is an open and inclusive community that recognizes community input according to the all-contributors acknowledgement schema. Ultimately, the goals of the community are not only to provide tools for using physiological data, but also to facilitate collection and dissemination of such data. The future of physiopy is a comprehensive suite of open-source tools for every step in the process from cleaning and preprocessing physiological recordings to using physiologically-based measures to improve neuroimaging and physiological research across research domains. Physiopy and its associated tools can be found at https://github.com/physiopy.Acknowledgements
We would like to thank the broader physiopy community for their work bringing knowledge and expertise for incorporating non-neural physiological data into neuroimaging research.References
1. Liu, T. T. Noise contributions to the fMRI signal: An overview. NeuroImage 143, 141–151 (2016).2. Kim, S.-G. & Ogawa, S. Biophysical and physiological origins of blood oxygenation level-dependent fMRI signals. J. Cereb. Blood Flow Metab. 32, 1188–1206 (2012).
3. Grady, C. L., Rieck, J. R., Nichol, D., Rodrigue, K. M. & Kennedy, K. M. Influence of sample size and analytic approach on stability and interpretation of brain-behavior correlations in task-related fMRI data. Hum. Brain Mapp. 42, 204–219 (2021).
4. Turner, B. O., Paul, E. J., Miller, M. B. & Barbey, A. K. Small sample sizes reduce the replicability of task-based fMRI studies. Commun. Biol. 1, 1–10 (2018).
5. Button, K. S. et al. Power failure: why small sample size undermines the reliability of neuroscience. Nat. Rev. Neurosci. 14, 365–376 (2013).
6. Gorgolewski, K. J. et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci. Data 3, 160044 (2016).
7. Daniel Alcalá et al. physiopy/phys2bids: BIDS formatting of physiological recordings. (Zenodo, 2020). doi:10.5281/zenodo.4312673.
Figures

Figure 1. Physiopy contributors. Contributions are recognized according to the all-contributors specification, see emoji key for details.
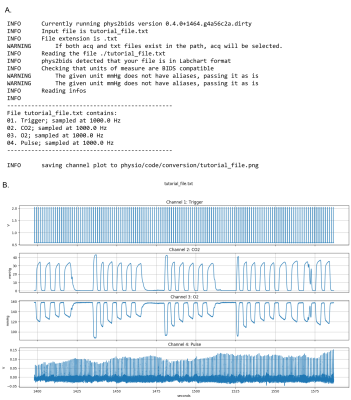
Figure 2. Using phys2bids to inspect the contents of physiological recording files. Choosing the ‘-info’ option in phys2bids allows users to inspect their data files before processing, providing information about recorded channels along with a graph.
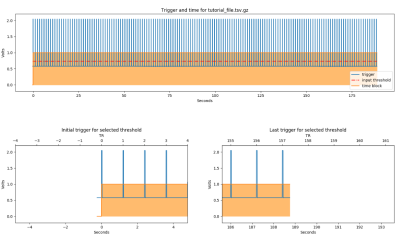
Figure 3. Trigger channel threshold estimation in phys2bids. In the minimal processing of phys2bids can rely on a “trigger channel” (A), indicating the duration of the (f)MRI acquisition(s) (or “runs”) (B, C), which phys2bids then uses to save recordings in BIDS-compliant, run-specific output files.
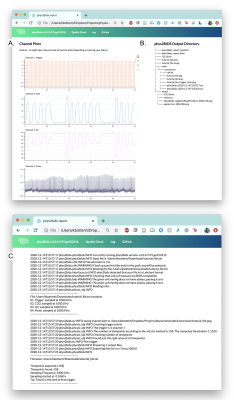
Figure 4. Reporting output of phys2bids. The phys2bids interface automatically creates an html report that includes (A) an interactive plot per channel that allows users to more closely inspect their data by panning and zooming along the time series, (B) a print-out of the output directory containing processed files, and (C) a log of the function call and subprocesses.