0725
QSMxT - A cross-platform, flexible, lightweight, and scalable processing pipeline for quantitative susceptibility mapping1Centre for Innovation in Biomedical Imaging Technology, University of Queensland, Brisbane, Australia, 2Centre for Advanced Imaging, University of Queensland, Brisbane, Australia, 3High Field MR Center, Department of Biomedical Imaging and Image-Guided Therapy, Medical University of Vienna, Vienna, Austria, 4Department of Neurology, Medical University of Graz, Graz, Austria, 5Siemens Healthcare Pty Ltd, Brisbane, Australia, 6Clinical & Research Imaging Centre, South Australian Health and Medical Research Institute, Adelaide, Australia, 7School of Information Technology and Electrical Engineering, University of Queensland, Brisbane, Australia
Synopsis
We developed an open-source QSM processing framework, QSMxT, that provides a full QSM workflow including converting DICOM data to BIDS, a variety of robust masking strategies, phase unwrapping, background field correction, dipole inversion and region-of-interest analyses based on automated anatomical segmentations. We make all required external dependencies available in a reproducible and scalable analysis environment enabling users to process QSM data for large groups of participants on any operating system in a robust way.
Introduction
Quantitative Susceptibility Mapping (QSM) is a post-processing technique applied to gradient-echo phase data. QSM processing involves a complex reconstruction pipeline requiring a series of processing steps including the conversion of DICOM data to a suitable analysis format, deriving masks to indicate reliable phase values for phase unwrapping, background field correction and dipole inversion. Finally, studies involving multiple participants require the construction of a common group space to facilitate group analyses and segmenting the resulting maps to extract quantitative values in anatomical regions of interest. Most QSM processing pipelines only focus on the unwrapping, background field correction and dipole inversion steps and neglect these pre- and post-processing tasks. Recent developments in the field1 aim to also include the masking procedure but rely on proprietary software (e.g. MATLAB) and are not easily applicable outside the human brain. We developed an open-source QSM processing pipeline, QSMxT, that automates all analysis steps from raw data preparation to region of interest analyses along with all required dependencies enabling users to process QSM data in a large group of participants. QSMxT is available on every operating system via a modular and containerised ecosystem, making our developments scalable and reproducible.Methods
We developed a modular reconstruction and analysis ecosystem that includes a variety of open source tools for computing and analysing QSM data. The first module of our ecosystem consists of a continuous integration system using GitHub actions to automatically build various software containers, including the containers required for our QSMxT framework. The necessary containers for QSMxT provide the FMRIB Software Library (FSL) v6.0.4, FreeSurfer (v7.1.1), Advanced Normalisation Tools (ANTs) v2.2.0, Julia v1.4.2, Bidscoin v.2.3, Python v3.8, TGV-QSM v1.0.0, the Python packages pydicom, nibabel, and Nipype, the Julia packages ArgParse and MriResearchTools, as well as our QSMxT NiPype workflow. QSMxT runs natively on Linux operating systems and on High-Performance clusters only requiring Singularity2 as a dependency. We also provide a lightweight Linux desktop conveniently accessible via a browser interface that runs on Windows and Mac only requiring docker as a dependency.Our QSMxT framework utilizes dicomsort, Bidscoin and dcm2niix to convert DICOM data into the BIDS standard3. For masking, we introduce two novel masking techniques designed to improve QSM results, especially in comparison to the commonly applied technique of generating a single brain mask for multi-echo data: First, our framework provides a new strategy that uses echo-dependent masks generated using the Brain Extraction Tool4 (BET) from the FSL, which reduces the influence of streaking artefacts near the brain boundary at later echo times. Second, it introduces a two-pass masking and QSM reconstruction strategy that processes subtle susceptibility sources independently from strong sources, reducing streaking artefacts, especially surrounding strong susceptibility sources such as veins and calcifications. This technique involves generating two masks for each echo, with the first mask based on a thresholded magnitude or phase-coherence, and the second step filling remaining gaps using morphological operations. This allows the processing of strong sources with rapid signal decay separately from subtle sources with longer T2*. The thresholding operation does not require anatomical priors, making it applicable to regions outside the brain, such as the abdomen or joints. The QSM inversion is performed using TGV-QSM5 with Laplacian unwrapping and combining background field correction and dipole inversion in a single optimization procedure. The resulting susceptibility maps are then averaged across echoes for each subject, and combined into group templates using minimum deformation averaging implemented in volgenmodel6. Image segmentation is performed using FreeSurfer7 on an MP(2)RAGE anatomy scan coregistered to the GRE data using ANTs, and scripts are provided to automatically extract susceptibility values from all participants in regions-of-interest to produce summary plots and statistics. Our data processing pipeline is visualised in terms of evolving file and folder structures in Figure 1, with the Nipype pipeline illustrated in terms of processing steps and brain images in Figure 2.
Results
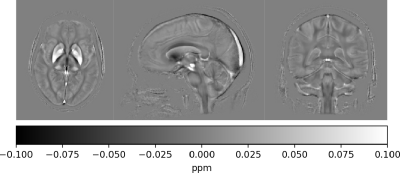
Our QSMxT framework provides a scalable and open workflow for QSM processing and analysis. The framework is implemented in a modular and reproducible ecosystem and here we show the processing of multi-echo QSM data for 45 gradient-echo brain acquisitions of healthy volunteers as an example. QSM templates based on the results from all subjects were automatically constructed and can be found in Figure 3, demonstrating the robustness and high quality of the QSM reconstructions obtained.Our framework does not require any installation or specific operating systems due to the use of container technology lowering the barriers for entry for less-technical users, while still being able to scale processing when required. QSMxT natively supports parallelization across CPU cores in one machine or distributed tasks in high-performance cluster batch systems such as Slurm or Grid Engine.
Conclusion
We developed a scalable and reproducible way of running a full QSM processing pipeline on any computer platform. Our QSMxT framework is fast and effortless to set up, relying mostly on point-and-click actions on the lightweight desktop environment. Our results demonstrate success in QSM applications at scale, as we have successfully automated the reconstruction process across a large multi-echo dataset on a cluster computing system. Our developments make QSM more accessible and overcome technical limitations that hamper its use and uptake.Acknowledgements
We thank the participants involved in this study. MB acknowledges funding from Australian Research Council Future Fellowship grant FT140100865 and SR from the Marie Skłodowska-Curie Action MS-fMRI-QSM 794298. This research was conducted by the Australian Research Council Training Centre for Innovation in Biomedical Imaging Technology (project number IC170100035) and funded by the Australian Government. Additional support was provided by the Austrian Science Fund (FWF): 31452 and KLI-646. The authors acknowledge the facilities and scientific and technical assistance of the National Imaging Facility, a National Collaborative Research Infrastructure Strategy (NCRIS) capability, at the Centre for Advanced Imaging, the University of Queensland. We wish to acknowledge QCIF for its support in this research by providing high-performance computing and storage resources.
Kieran O’Brien and Jin Jin are employees of Siemens Healthineers in Australia. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationship that could be construed as a conflict of interest.
References
[1] Chan, Kwok-Shing, Marques, José P., 'SEPIA - SuscEptibility mapping PIpeline tool for phAse images,' Neuroscience, preprint, Jul. 2020. https://doi.org/10.1101/2020.07.23.217042.
[2] Kurtzer, Gregory M., Vanessa Sochat, and Michael W. Bauer. 'Singularity: Scientific Containers for Mobility of Compute'. PLOS ONE 12, no. 5 (11 May 2017): e0177459. https://doi.org/10.1371/journal.pone.0177459
[3] Gorgolewski, Krzysztof J., Tibor Auer, Vince D. Calhoun, R. Cameron Craddock, Samir Das, Eugene P. Duff, Guillaume Flandin, et al. 'The Brain Imaging Data Structure, a Format for Organizing and Describing Outputs of Neuroimaging Experiments'. Scientific Data 3 (21 June 2016): 160044. https://doi.org/10.1038/sdata.2016.44
[4] S. M. Smith, 'Fast robust automated brain extraction,' Human Brain Mapping, vol. 17, no. 3, pp. 143–155, 2002. 10.1002/hbm.10062
[5] Langkammer, Christian, Kristian Bredies, Benedikt A. Poser, Markus Barth, Gernot Reishofer, Audrey Peiwen Fan, Berkin Bilgic, Franz Fazekas, Caterina Mainero, and Stefan Ropele. 'Fast Quantitative Susceptibility Mapping Using 3D EPI and Total Generalized Variation'. NeuroImage 111 (1 May 2015): 622–30. https://doi.org/10.1016/j.neuroimage.2015.02.041
[6] Janke, Andrew L., and Jeremy F. P. Ullmann. 'Robust Methods to Create Ex Vivo Minimum Deformation Atlases for Brain Mapping'. Methods, Spatial mapping of multi-modal data in neuroscience, 73 (February 2015): 18–26. https://doi.org/10.1016/j.ymeth.2015.01.005
[7] Fischl, B., Salat, D. H., Busa, E., Albert, M., Dieterich, M., Haselgrove, C., ... & Montillo, A. 'Automated Labeling of Neuroanatomical Structures in the Human Brain'. Neuron, (2002): 33:341-355.
Figures