0714
Automatic fetal ocular measurements in MRI1School of computer science and engineering, Hebrew University of Jerusalem, Jerusalem, Israel, 2Sagol Brain Institute, Tel Aviv Sourasky Medical Center, Tel Aviv, Israel, 3Sagol School of Neuroscience, Tel Aviv University, Tel Aviv, Israel, 4Division of Pediatric Radiology, Tel Aviv Sourasky Medical Center, Tel Aviv, Israel, 5Sackler Faculty of Medicine, Tel Aviv University, Tel Aviv, Israel, 6Medical Imaging, Children's Hospital of Eastern Ontario, University of Ottawa, Ottawa, ON, Canada
Synopsis
The aim of this study was to establish a fully automatic method for ocular measurements from in-vivo fetal brain MRI. Axial brain MRI of 47 fetuses (29-38 weeks’ gestational age) were included. The method includes fetal brain ROI computation and fetal eye segmentation using deep learning, followed by geometric algorithms for 2D and 3D measurements of the binocular (BOD), interocular (IOD), and ocular (OD) diameters. The performance of the 2D measurements was found to be preferable over 3D, with <1mm deviation from manual expert neuro-radiologist annotations. This is the first fully automatic method for fetal ocular biometric measurements in MRI.
Introduction
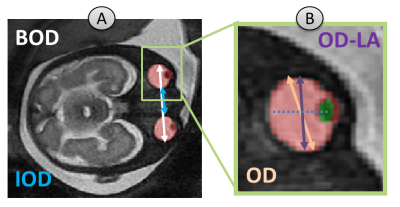
Fetal ocular biometrics are important parameters for fetal growth evaluation and detection of congenital abnormalities during pregnancy such as hypertelorism, hypotelorism, microophtalmia and anophthalmia, as they can be part of a genetic syndrome or may be related to a developmental abnormality. Accurate measurements can support improved diagnosis, pregnancy and birth management. Ocular biometrics including binocular (BOD), interocular (IOD), and ocular (OD) diameters (Figure 1) are manually measured in routine clinical practice, thus are dependent on the annotator’s expertise. A few studies proposed methods for ocular measurements, including manual measurements proposed by Robinson1 and Li2, which requires the OD to be perpendicular to the eye lens. A semi-automatic algorithm for 3D measurements was suggested by Velasco-Annis3 based on motion corrected reconstructed volumes.The aim of this study was to develop fully automatic methods for ocular measurements from in-vivo fetal brain MRI, based on the previously suggested methods.
Methods
The method consists of four stages (Figure 2): (1) fetal brain ROI computation; (2) fetal orbit segmentation; (3) lens and globe segmentation; and (4) ocular measurements.Subject data: Axial brain MRI volumes of 47 fetuses (gestational age: 29-38 weeks) were included in this study. Scans were performed on 1.5T General Electric or 3T Siemens systems and included T2-weighted FRFSE or HASTE sequences, with in-plane resolution of 0.45-1.25mm and slice thickness of 2-5mm.
Data annotation and splitting: Manual annotations of BOD, IOD and OD measurements and orbits, lens and globe segmentations were performed by a senior pediatric neuro-radiologist. For the segmentation stages, the dataset was split into 16 training, 4 validation and 27 test volumes.
Fetal brain ROI computation: The fetal brain was first segmented using a 3D two-stage anisotropic U-Net4, and then cropped using axis-aligned 3D bounding box of the resulting segmentation.
Fetal orbit, lens, and globe segmentation: Deep learning models were developed using the Fast.ai framework5. A 2D U-Net6 with pre-trained ResNet347 convolutional neural network as encoder network was used. Network training was performed with Dice loss8 and batch size of 8 slices. The network was trained for 24 epochs with OneCycle scheduler9 with initial learning rate of 1e-3.
Due to the relatively small annotated dataset size, the network was trained using a pre-trained encoder model based on ResNet34 ImageNet10 pre-trained weights. In addition, data augmentations of 2D rotation, brightness and contrast adjustment were performed. Two models were trained, one for orbit segmentation, and a second one for lens and globe segmentation.
The orbit segmentation results are clustered into the two largest connected components, corresponding to the fetus’ two eyes. Similarly, for the lens and globe model, the two largest clusters, each one composed of lens and globe voxels, were selected.
Ocular Measurement algorithm: All measurements were automatically computed. BOD-2D was computed as the maximum distance found between any two voxels between the two orbits, on all slices. IOD-2D was computed as the minimum distance found between two voxels between the two orbits, on all slices. OD-2D was computed as the maximum distance between any two voxels within a single orbit (for both eyes), on all slices. These 2D measurements were acquired in a similar manner to Robinson1. OD-LA-2D (Lens Aligned) was computed as the maximum diameter of the globe boundary voxels perpendicular to the line between globe center and lens center of the eye, similar to Li2. BOD-3D, IOD-3D and OD-3D were calculated similar to our 2D measurements, but on the total orbit volume, similar to Velasco-Annis3. In this study, OD of any type (OD-2D, OD-LA-2D and OD-3D) is reported as the mean diameter of the two eyes.
Evaluation of results: 3D Dice coefficient was used to quantify the performance of the automatic segmentation results compared to manual segmentations. Estimates of mean difference and Bland–Altman plots were used to quantify differences between the automatic and manual ocular measurements.
Results
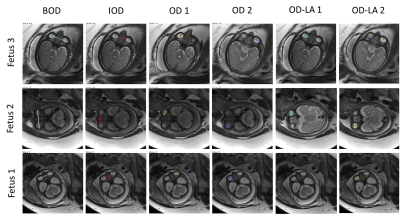
Segmentation results: Automatic orbit segmentation achieved a mean 3D Dice of 92.9% (89.0-95.0), and lens and globe segmentation achieved a mean Dice of 93.7% (91.5-95.2).Measurement results: Table 1 presents the Bland-Altman (bias and agreement) and mean difference metrics for each measurement compared to manual annotations. As can be observed, the 2D measurements were more accurate than the 3D, with <1mm mean difference and <2mm variability compared to manual annotation. Figure 3 shows representative examples of automatic 2D ocular measurements from three fetuses.
Discussion and Conclusions
This work presents the first fully automatic method for computing fetal ocular biometric measurements from fetal brain MRI. Following segmentation, measurements were obtained using different methods: 2D, 3D and lens aligned (OD-LA-2D). Automatic 2D measurements may be preferable compared to 3D, as 3D reconstruction may be affected by fetal movements and partial volume effects, caused by the anisotropy acquired resolution. The highest performance of OD was achieved using OD-LA-2D, as it is similar to the clinical guidelines of OD measurements used in this study for annotation. However, future studies on a larger cohort should explore the sensitivity and variability of each method, to achieve optimization for clinical assessment. To conclude, this method is accurate, scanner independent and reproducible, and thus has the potential to be integrated in routine clinical setup.Acknowledgements
This work was supported by Kamin grants of the Israel Innovation Authority.References
1. Robinson, A.J., et al., MRI of the fetal eyes: morphologic and biometric assessment for abnormal development with ultrasonographic and clinicopathologic correlation. Pediatric Radiology, 2008. 38(9): p. 971-981.
2. Li, X.B., et al., Fetal ocular measurements by MRI. Prenatal diagnosis, 2010. 30(11): p. 1064-1071.
3. Velasco‐Annis, C., et al., Normative biometrics for fetal ocular growth using volumetric MRI reconstruction. Prenatal diagnosis, 2015. 35(4): p. 400-408.
4. Dudovitch, G., et al. Deep Learning Automatic Fetal Structures Segmentation in MRI Scans with Few Annotated Datasets. in Proc. International Conference on Medical Image Computing and Computer-Assisted Intervention. 2020.
5. Howard, J. and S. Gugger, Fastai: A Layered API for Deep Learning. Information, 2020. 11(2): p. 108.
6. Ronneberger, O., P. Fischer, and T. Brox. U-net: Convolutional networks for biomedical image segmentation. in in Proc. International Conference on Medical Image Computing and Computer-Assisted Intervention. 2015.
7. He, K., et al. Deep residual learning for image recognition. in Proc. of the IEEE conference on Computer Vision and Pattern Recognition. 2016.
8. Sudre, C.H., et al., Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations, in Deep Learning in Medical Image Analysis and multimodal learning for clinical decision support. 2017.
9. Smith, L.N., A disciplined approach to neural network hyper-parameters: Part 1--learning rate, batch size, momentum, and weight decay. arXiv preprint arXiv:1803.09820, 2018.
10. Deng, J., et al. Imagenet:
A large-scale hierarchical image database. in Proc. of the IEEE conference on Computer Vision and Pattern Recognition.
2009.
Figures