0629
Highly Accelerated Sub-millimeter Resolution 3D EPI using Variable Density CAIPI Sampling with Temporal Random Walk for Functional MRI at 7 Tesla1Department of Computer Engineering, Chonnam National University, Gwangju, Korea, Republic of, 2Department of ICT Convergence System Engineering, Chonnam National University, Gwangju, Korea, Republic of, 3Siemens-Healthineers, Seoul, Korea, Republic of, 4Center for Neuroscience Imaging Research (CNIR), Institute for Basic Science (IBS), Suwon, Korea, Republic of, 5Department of Biomedical Engineering, Sungkyunkwan University, Suwon, Korea, Republic of, 6University of California, Berkeley, Berkeley, CA, United States, 7Advanced MRI Technologies, Sebastopol, CA, United States
Synopsis
With ultra-high fields, 3D EPI has been used by improving imaging efficiency. Nevertheless, there have been some limitations: 1) the regular sampling limits the use of temporal structure in the data and 2) parallel imaging allows up to 6-fold acceleration in 3D acquisition. Here, we developed an accelerated 3D EPI using VD-CAPI sampling with temporal random walk. Experimental studies confirm advantages in acceleration, SNR, and sensitivity of the proposed method: 1) temporal random walk allows extra spatial encoding across time, 2) temporal prior provides high SNR, and 3) the temporal incoherent sampling and high SNR result in higher BOLD activations.
Introduction
With ultra-high magnetic fields, functional MRI (fMRI) achieves increased signal-to-noise ratio (SNR) and sensitivity to BOLD contrast at sub-millimeter resolutions, and hence holds great potential for columnar and layer imaging1-4. In pursuit of sub-millimeter resolution fMRI with a short TR per volume, application of 2D CAIPI sampling to multi-shot 3D EPI was recently proposed5-7, improving acquisition efficiency with reduced g-factor penalty during reconstruction. Nevertheless, 3D EPI acquisitions have some disadvantages in that 1) the regular sampling with the EPI sequence limits the use of temporal structure in the data and 2) parallel imaging typically allows up to 6-fold acceleration in 3D acquisition even with 32 receiver channels8-9. In this work, we developed a novel highly accelerated 3D EPI using variable density (VD) 2D CAPI sampling with temporal random walk. Experimental studies confirm advantages in acceleration, SNR, and sensitivity of the proposed new sampling scheme coupled with 3D EPI: 1) temporal random walk in 3D EPI allows extra spatial encoding by exploiting the temporal redundancy, 2) temporally constrained reconstruction provides high SNR efficiency, and 3) the incoherent time-varying sampling and high SNR efficiency result in higher BOLD sensitivity in the expected primary visual areas.Methods
3D Spatiotemporal Encoding: The design of the spatiotemporal sampling in 3D EPI is important in estimating missing signals from highly undersampled data. In order to design an optimized random encoding for fMRI, we introduce a temporal random walk into 2D VD-CAIPI sampling under the framework of 3D EPI sequence. Fig. 1A shows an example of the temporal random walk. For the spatial encoding, CAIPI-based sampling repeats for all TRs by evenly distributing the aliasing between both of the phase encoding axes. In each TR, the CAIPI sampling itself randomly jumps to neighboring sites of the lattice within a ky -kz block (the bock size is the undersmapled factor for each axis) to achieve a complementary random encoding across time while keeping the coherence on the spatial axes. Additionally, we incorporate the VD concept along the partition axis into 2D CAIPI sampling to include more central k-space data without changing EPI blip size along phase axis (as shown in Fig. 1B).Reconstruction: For spatiotemporal joint reconstruction, SENSE is applied to the spatial axes while low rank and sparsity priors are applied to temporal axis under the assumption that parallel imaging is more effective with the regular sampling while the above temporal priors well performed with incoherent acquisition10-11.
Experiments: All experiments were conducted using a 7T whole-body scanner (MAGNETOM, Terra, Siemens Healthineers, Erlangen, Germany) equipped with a 32-channel head coil. Institutional review board and informed consent was obtained for all subjects. All data were acquired using accelerated 3D EPI with 1) Skipped-CAIPI and 2) VD-CAIPI+Random Walk. The imaging parameters are summarized in table 1.
Stimulation paradigm: Functional activation was assessed by performing a flickering checkerboard pattern with 6.25 Hz frequency in blocks of 19.2s on/off for visual stimulation. The cycle was repeated 8 times per scan for a total task duration of 5.44 minutes.
Results
Fig. 2 shows an example of reconstructed images (displaying 6 out of 60 slices) acquired using skipped CAIPI (6-fold acceleration) and proposed VD-CAIPI+Random Walk (8-fold acceleration). The skipped CAIPI yields aliasing in frontal area and signal loss around slab boundary. On the other hand, the proposed method substantially suppresses image noises and aliasing artifacts over the entire brain. Fig. 3 and 4 compares functional activation maps of primary visual cortex using multiplanar reformatting for each sequence. Note that skipped CAIPI introduces a smearing of BOLD information, while the proposed method can better delineate cortical gray matter area with higher BOLD sensitivity.Conclusions
We demonstrated that multi-shot 3D EPI combined with a new sampling scheme (VD-CAIPI+Random Walk) has significant improvement over the current skipped CAIPI 3D EPI sequence in terms of net acceleration, reconstruction quality, and functional activation. Unlike the current sequence that operates in a time-independent manner for reconstruction, the proposed method is able to exploit temporal redundancy while keeping normal EPI framework unchanged, thus improving statistical power in measuring functional activation. In the near future, we plan to incorporate the new 3D sequence readout with CBV-based VASO sequences12.Acknowledgements
This work has been supported by the National Institute of Biomedical Imaging and Bio-engineering, and the National Institute for Mental Health of the National Institutes of Health and the BRAIN Initiative under award numbers: U01EB025162 and 1R01MH111444References
1. Huber L, Ivanov D, Handwerker DA, Marrett S, Guidi M, Uludağ K, Bandettini PA, Poser BA. Techniques for blood volume fMRI with VASO: From low-resolution mapping towards sub-millimeter layer-dependent applications. Neuroimage. 2018; 164:131-143.
2. Goense J, Merkle H, Logothetis NK. High-resolution fMRI reveals laminar differences in neurovascular coupling between positive and negative BOLD responses. Neuron. 2012; 76(3):629-639.
3. Kashyap S, Ivanov D, Havlicek M, Sengupta S, Poser BA, Uludağ K. Resolving laminar activation in human V1 using ultra-high spatial resolution fMRI at 7T. Scientific reports. 2018; 8(1):1-1.
4. Park S, Torrisi S, Townsend JD, Beckett A, Feinberg DA. Highly accelerated submillimeter resolution 3D GRASE with controlled blurring in‐weighted functional MRI at 7 Tesla: A feasibility study. Magn Reson Med. 2020; doi:https://doi.org/10.1002/mrm.28589
5. Poser BA, Koopmans PJ, Witzel T, Wald LL, Barth M. Three dimensional echo-planar imaging at 7 Tesla. Neuroimage. 2010 May 15;51(1):261-266.
6. Le Ster C, Moreno A, Mauconduit F, Gras V, Stirnberg R, Poser BA, Vignaud A, Eger E, Dehaene S, Meyniel F, Boulant N. Comparison of SMS-EPI and 3D-EPI at 7T in an fMRI localizer study with matched spatiotemporal resolution and homogenized excitation profiles. PLoS One. 2019; 14(11): e0225286.
7. Stirnberg R, Stöcker T. Segmented K‐space blipped‐controlled aliasing in parallel imaging for high spatiotemporal resolution EPI. Magn Reson Med. 2020; doi:https://doi.org/10.1002/mrm.28486.
8. Breuer FA, Blaimer M, Mueller MF, Seiberlich N, Heidemann RM, Griswold MA, Jakob PM. Controlled aliasing in volumetric parallel imaging (2D CAIPIRINHA). Magn Reson Med. 2006; 55(3):549-556.
9. Brenner D, Stirnberg R, Pracht ED, Stöcker T. Two-dimensional accelerated MP-RAGE imaging with flexible linear reordering. Magn Reson Mate Physics Biol Med. 2014; 27(5):455-462.
10. Akçakaya M, Basha TA, Chan RH, Manning WJ, Nezafat R. Accelerated isotropic sub‐millimeter whole‐heart coronary MRI: compressed sensing versus parallel imaging. Magnetic resonance in medicine. 2014; 71(2):815-822.
11. Miao J, Guo W, Narayan S, Wilson DL. A simple application of compressed sensing to further accelerate partially parallel imaging. Magn Reson Imaging. 2013; 31(1):75-85.
12. Huber L, Chai Y, Stirnberg R, Kashyap S, Kurban D, Khojandi A, Ivanov D, Marrett S, Stöcker T, Uludag K, Bandettini P. Beyond the limits of layer-dependent CBV fMRI in humans: strategies towards whole brain coverage, sub-second TR, and very high 0.5 mm resolutions. In Proceedings of the international society of magnetic resonance in medicine 2020; p.3864.
Figures
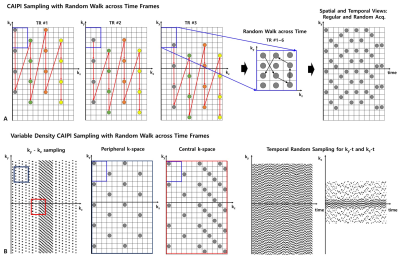
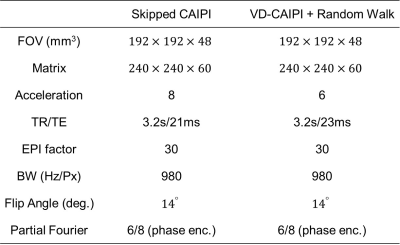
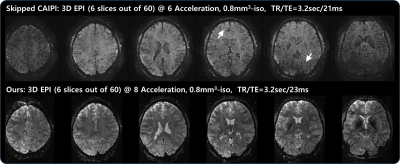
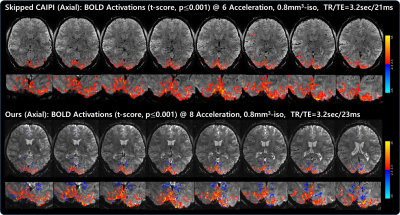
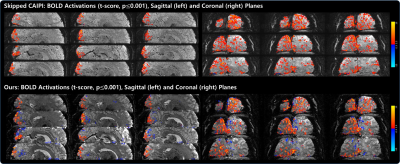
Fig. 4. Comparisons of visual activation maps (t-score, p≤0.001) overlaid on the average 3D EPI images observed from both sagittal (left) and coronal (right) views: skipped CAIPI (top) vs.VD-CAIPI+Random Walk (bottom). Consistent with the axial results shown in Fig. 3, the proposed method can better delineate cortical GM area with higher BOLD sensitivity.