0607
Deep Learning-based high-resolution pseudo-CT to detect cranial bone abnormalities for pediatric patients using MRI1Mallinckrodt Institute of Radiology, Washington University in St. Louis, Saint louis, MO, United States, 2Department of Neurology, Washington University in St. Louis, Saint louis, MO, United States, 3Division of Plastic and Reconstructive Surgery, Washington University in St. Louis, Saint louis, MO, United States
Synopsis
Computed tomography (CT) scans are commonly used in pediatric patients with head trauma and craniosynostosis to identify skull fractures and sutures, respectively. However, the ionizing radiation associated with the CT scans increases the pediatric patients’ risk for cancer. We developed a deep learning-based method, which consists of two networks focusing on skull and head separately, to generate high-resolution pseudo-CT (pCT) from a radial MR scan. A Dice coefficient of 0.90 ± 0.02 was obtained in the bone.Moreover, a pCT mean absolute error (MAE) of 87.5 ± 4.4 HU was achieved.
Introduction
Head trauma is common in the pediatric population resulting in approximately 600,000 emergency cases per year.1 On the other hand, craniosynostosis is the abnormal early fusion of a cranial suture, causing an irregularly shaped cranium.2 3D high-resolution head CT scans are commonly used in these pediatric patients to identify skull fractures and sutures. However, the ionizing radiation associated with the CT scans increases the risk of cancer. The National Cancer Institute reported that radiation exposure from multiple head CT scans in children might triple the risk of leukemia and brain cancer.3-8 Magnetic Resonance Imaging (MRI) does not use ionizing radiation and can be a safe alternative to CT. A gradient-echo “Black Bone” (BB) MR sequence and an ultra-short TE MR sequence with pointwise encoding and radial acquisition were previously proposed but suffer from motion and sub-optimal contrast between bone and the surrounding tissues.9,10 A time-consuming and subjective manual post-processing methods need to be performed. In this study, we implemented a golden angle (GA) stack-of-stars radial MR sequence and developed a deep learning (DL) model to generate 3D high-resolution cranial bone pseudo-CT (pCT) images from MR GA images. The DL model is robust and fully automated to facilitate translation to clinical use.Methods
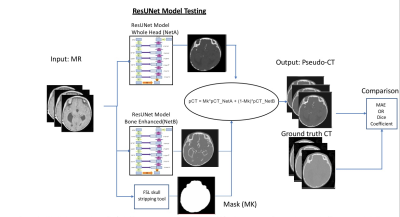
After obtaining IRB approval, St. Louis Children’s hospital pediatric patients (ages three days to 11 years old) diagnosed with either head trauma or craniosynostosis were recruited for this study. Participants had a standard head clinical CT scan and a research MR GA scan.11 The MR GA paramaters were 224 slices, TE/TR = 2.47ms/4.84ms, Flip angle = 3°, BW = 410 Hz/pixel, FOV = 192 mm, Voxel size 0.6x0.6x0.8 mm, Number of radial spokes = 400, Total acquisition time = 5:04. Image preprocessing included N4 bias field correction and an affine image registration between MR and CT images using FSL transformation with a bone weighted cost function.12,13 A BM3D filter (edge-preserving smoothing) was applied to CT images.14 We trained two different Residual UNet (ResUNet) networks (Figure 1). For the first network, all patches were selected randomly from the whole head. The ResUNet model utilized over one million 3D partially overlapping patches (size 64x64x64 voxels) to increase the number of model training samples.15-17 Since cranial bone and sutures only occupy a small fraction of the total imaging volume, and they are not represented well in the training samples in the first ResUNet with randomly selected patches. To better identify the skull and sutures, we enhanced their representation by increased sampling patches near the skull and sutures in the second ResUNet. Both networks were implemented using Pytorch libraries, and the weights were updated by reducing the L1 loss function difference between the pCT output and the target CT image with an Adam optimizer using a learning rate parameter of 0.001 and a batch size of 10. Figure 2 demonstrated that the skull enhanced ResUNet converged after 50,000 iterations. The final pCT was generated by combining the outputs from both networks, as detailed in Figure 3. The quality of the combined pCT images was evaluated using the Dice coefficient and the mean absolute error (MAE) with respect to the acquired CT images. Given the small sample size of the study, a leave-one-out cross-validation was performed with 16 patients in training and the remaining one patient in testing. This leave-one-out cross-validation was repeated 12 times to obtain testing results from 12 patients. In order to assess the deep learning-based pCT results, MR images were manually processed by an experienced operator. The images were bias-field corrected, followed by an intensity inversion, and a threshold was applied to separate bone from other tissues. The manual data processing took approximately 30 minutes to 2 hours per subject. The DL method, however, was fully automatic and fast for subject testing (a few minutes per subject).Results
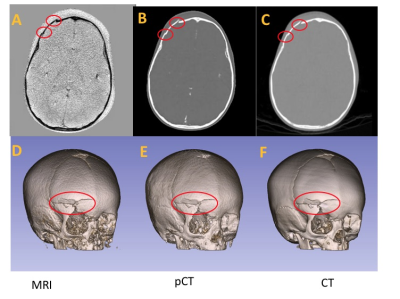
The DL methods demonstrated CT-equivalent 3D cranial bone images from pediatric subjects can be created using a GA MR scan. Figure 4 displays sample pCT and CT images for visualizing sutures in four participants , and Figure 5 shows an example from a trauma patient with fractures.The proposed deep learning ResUNet models generated pCT images that closely resemble the gold-standard CT images. The MAE between pCT and CT was 87.5 ± 4.4 HU. The mean and standard deviation of the Dice coefficients were 0.90 ± 0.02. The bone Dice coefficients were 0.76 ± 0.07 from six patients using a manually determined participant-specific threshold for bone segmentation. In comparison, the Dice coefficients between the deep learning pCT and CT bones were 0.89 ± 0.02 for the same six patients. Our deep learning method achieved a significantly higher Dice coefficient than the manual processing scheme (p= 0.0016).Discussion and Conclusions
A robust and fully automated DL method to convert MRI images into pCT can facilitate translating MR cranial bone imaging into clinical practice for pediatric patients. A Golden Angle stack-of-stars scan can provide high-resolution pCT images. The proposed DL method outperformed a manual post-processing method in achieving high Dice coefficients. The DL model performance may depend upon the age range of participants. In the future, we will continue to investigate the age dependence of this method.Acknowledgements
No acknowledgement found.References
1. Schneier AJ, Shields BJ, Hostetler SG, Xiang H, Smith GA. Incidence of pediatric traumatic brain injury and associated hospital resource utilization in the United States. Pediatrics2006; 118(2): 483-92.
2. French LR, Jackson IT, Melton LJ, 3rd. A population-based study of craniosynostosis. J Clin Epidemiol1990; 43(1): 69-73.
3. Dorfman AL, Fazel R, Einstein AJ, Applegate KE, Krumholz HM, Wang Y, Christodoulou E, Chen J, Sanchez R, Nallamothu BK. Use of medical imaging procedures with ionizing radiation in children: a population-based study. Arch Pediatr Adolesc Med. 2011;165(5):458-64. Epub 2011/01/05. doi: 10.1001/archpediatrics.2010.270. PubMed PMID: 21199972; PMCID: PMC3686496.
4. Miglioretti DL, Johnson E, Williams A, Greenlee RT, Weinmann S, Solberg LI, Feigelson HS, Roblin D, Flynn MJ, Vanneman N, Smith-Bindman R. The use of computed tomography in pediatrics and the associated radiation exposure and estimated cancer risk. JAMA Pediatr. 2013;167(8):700-7. Epub 2013/06/12. doi: 10.1001/jamapediatrics.2013.311. PubMed PMID: 23754213; PMCID: PMC3936795.
5. Brenner DJ, Elliston CD, Hall EJ, Berdon WE. Estimates of the cancer risks from pediatric CT radiation are not merely theoretical: comment on "point/counterpoint: in x-ray computed tomography, technique factors should be selected appropriate to patient size. against the proposition". Med Phys. 2001;28(11):2387-8. PubMed PMID: 11764047.
6. Parker L. Computed tomography scanning in children: radiation risks. Pediatr Hematol Oncol. 2001;18(5):307-8. doi: 10.1080/088800101300312564. PubMed PMID: 11452401.
7. Pearce MS, Salotti JA, Little MP, McHugh K, Lee C, Kim KP, Howe NL, Ronckers CM, Rajaraman P, Sir Craft AW, Parker L, Berrington de Gonzalez A. Radiation exposure from CT scans in childhood and subsequent risk of leukaemia and brain tumours: a retrospective cohort study. Lancet. 2012;380(9840):499-505. doi: 10.1016/S0140-6736(12)60815-0. PubMed PMID: 22681860; PMCID: 3418594.
8. Smyth MD, Narayan P, Tubbs RS, Leonard JR, Park TS, Loukas M, Grabb PA. Cumulative diagnostic radiation exposure in children with ventriculoperitoneal shunts: a review. Childs Nerv Syst. 2008;24(4):493-7. doi: 10.1007/s00381-007-0560-x. PubMed PMID: 18180935.
9. Eley KA, McIntyre AG, Watt-Smith SR, Golding SJ. "Black bone" MRI: a partial flip angle technique for radiation reduction in craniofacial imaging. Br J Radiol2012; 85(1011): 272-8.
10. Kralik SF, Supakul N, Wu IC, et al. Black bone MRI with 3D reconstruction for the detection of skull fractures in children with suspected abusive head trauma. Neuroradiology2019; 61(1): 81-7.
11. Patel KB, Eldeniz C, Skolnick GB, Jammalamadaka U, Commean PK, Goyal MS, Smyth MD, An H. 3D pediatric cranial bone imaging using high-resolution MRI for visualizing cranial sutures: a pilot study. J Neurosurg Pediatr. 2020 Jun 12:1-7. doi: 10.3171/2020.4.PEDS20131. Epub ahead of print. PMID: 32534502; PMCID: PMC7736460.
12. Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010;29(6):1310-20. doi: 10.1109/TMI.2010.2046908. PubMed PMID: 20378467; PMCID: PMC3071855.
13. Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17(2):825-41. PubMed PMID: 12377157.
14. Dabov Kostadi, Foi, Alessandro; Katkovnik, Vladimir; Egiazarian, Karen. Image denoising by sparse 3D transform-domain collaborative filtering. IEEE Transactions on Image Processing. 16 July 2007. 16 (8): 2080–2095
15. Ronneberger O, Fisher P, Brox T. U-Net: Convolutional Networks for Biomedical Image Segmentation Medical Image Computing and Computer-Assisted Intervention (MICCAI); 2015. p. 234-41.
16. He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. IEEE Computer Vision and Pattern Recognition (CVPR)2016. p. 770-8.
17. Ying C, Chen Y, Binkley M, Juttukonda MR, Flores S, Benzinger TLS, An H, editors. Deep learning based T1-enhanced selection of linear attenuation coefficients for PET/MR attenuation correction: accuracy and repeatability. International Society of Magnetic Resonance in Medicine; 2020.
Figures