0604
Disability Prediction in Multiple Sclerosis using Ensemble of Machine Learning Models and DTI Brain Connectivity1CREATIS (UMR 5220 CNRS & U1206 INSERM), Université Claude Bernard Lyon 1, Villeurbanne, France, 2Department of Mathematics and Computer Science, University of Calabria, Rende, Italy, 3R&D Department, CGnal, Milan, Italy, 4Hôpital Neurologique, Hospices Civils de Lyon, Bron, France, 5MRI, CERMEP - Imagerie du Vivant, Bron, France
Synopsis
The Expanded Disability Status Scale (EDSS) monitors physical impairment in Multiple Sclerosis (MS). A Staking Ensemble model composed of 4 ML "boosting" models was used to predict EDSS using both white matter (WM) fiber-bundles and structural connectome data. This model provided excellent prediction results with an RMSE of 0.92 to 1.08. A counterfactual model was added to highlight the most important WM links and fiber-bundles in the prediction process. The accordance of the findings obtained with both data types confirmed the clinical interest of such methods for disability prediction using DTI data.
Background or Purpose
Multiple sclerosis (MS) is an autoimmune inflammatory disease characterized by demyelination and neurodegeneration processes and leading to cognitive and physical impairments. In clinical practice, disability is mainly monitored using the Expanded Disability Status Scale (EDSS), which represents motor impairment. Diffusion tensor imaging (DTI) constitutes a very sensitive technique to detect white matter (WM) microstructural damages and to provide network graphs of the brain structural connectivity based on fiber-tracking1. The goal of this work is to develop first, an ensemble machine learning (ML) model to predict disability based on structural connectivity and WM fiber-bundles, and second, a novel interpretability model to better understand the WM networks underlying the ML prediction pathway.Methods
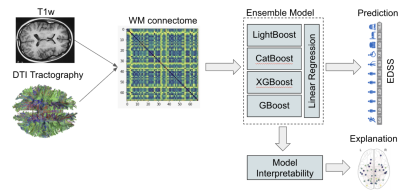
A Staking Ensemble composed of four ML "boosting" models, linked together by linear regression (Figure 1), was trained to predict MS patient's EDSS score based on 528 MRI exam of 114 MS patients divided into 4 clinical profiles: Clinical Isolated Syndrom (CIS), Relapsing-Remitting (RRMS), Primary Progressive (PP) and Secondary Progressive (SP).The MRI protocol was composed of a 3D T1-weighted (magnetization prepared rapid gradient echo-MPRAGE) sequence with repetition time/echo time/time for inversion (TR/TE/TI) = 1970/3.93/1100 ms, flip angle = 15°, matrix size = 2.5×2.5×2.5 mm3 field of view (FOV) = 256×256 mm, slice thickness = 1 mm, voxel size = 1×1×1 , acquisition time = 4.62 min and an axial 2D-spin-echo DTI sequence (TE/TR = 86/6900 ms; 2×24 directions of gradient diffusion; b = 1000 s.mm-2 , spatial resolution of 2.5×2.5×2.5 mm3 ) oriented in the AC-PC plane.After labeling each voxel of the T1w-MR images in four classes [WM, cortical GM, sub-cortical GM, cerebro-spinal fluid (CSF)], a parcellation of the cortical and sub-cortical GM was performed on the T1w images using the Desikan-Killiany2. DTI acquisitions were first processed using probabilistic streamline fiber-tracking algorithm to generate WM fibers, which were used as connections between the GM nodes, leading to the construction of a symmetrical connectivity matrix A ∈ N+qxq where q=84 for each patient for the connectome data. Second, diffusion maps of each subject were co-registered (non-rigid) on the Illinois Institute of Technology3 (IIT3) atlas and 20 fiber-bundles were extracted using the JHU4 atlas.
The Ensemble model was applied to both structural connectivity data (matrices) and WM fiber-bundles for the estimation of disability. In addition, an interpretability model, based on counterfactual perturbation, was developed to explain the global and local behaviors of the Ensemble model.
Results
Both approaches provided excellent prediction results with an RMSE of 0.92 (± 0.28) using the connectome data (Figure 2) and an RMSE of 1.08 (± 0.12) using the fiber-bundles data.The two approaches were compared using a non-parametric Mann-Whitney U test and no significant difference was obtained (p = 0.48). Depending on the classes of disability (Low : 0-3, medium: 3-4.5, high: 4.5), the highest prediction error was found in patients with low disability [0-3] while the lowest was found in patients with medium disability [3-4.5] (Figure 3).
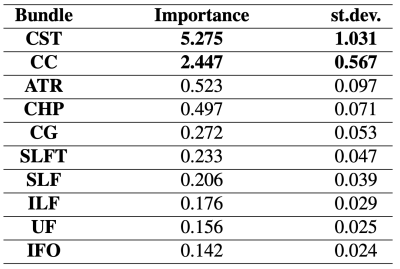
The interpretability model highlighted the brain networks connecting frontal, temporal and parietal regions as the most important links5 for predicting disability when connectome data were used (Figure 4), as previously observed in MS6,7. More in detail, left-hemispheric as well as inter-hemispheric connections (Corpus Callosum (CC))8 were found to be the most relevant. Interestingly, when using WM fiber-bundles, the CC was again considered as well as the Cortical Spinal Tract (CST), which is related to the physical impairment measured by EDSS (Table 1).
Conclusion
In this study, we demonstrated that it is possible to predict the EDSS score with great accuracy. A new interpretability model was proposed in order to better understand the brain networks deemed relevant by the model for predicting disability. The two approaches based on different data (WM connectome and fiber-bundles) provided a similar degree of accuracy and highlighted similar WM fiber-tracts when the interpretability model was applied. These preliminary findings increased our confidence in the proposed models and confirmed the clinical interest of such DTI based approaches.Acknowledgements
This study is funded by the following projects: European Research Council within the grant 813120-2018 of Marie Sklodowska-Curie Innovative Training Networks (ITN) of the Horizon 2020 through the INSPiRE-MED project. French National Research Agency (ANR) within the national program “Investissements d’Avenir” through the OFSEP project (ANR-10-COHO-002).References
1. Rovira A., Auger C. et al., “Magnetic resonance monitoring of lesion evolution in multiple sclerosis,” Ther Adv Neurol Disord, 6(5):298–310:2013.
2. Desikan R.S., Ségonne F., Fischl B., et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage. 31(3):968-80:2006.
3. Varentsova A., Zhang S. et al., “Development of a high angular resolution diffusion imaging human brain template,” Neuroimage, 91:177–186:2014.
4. K. Hua, J. Zhang et al., “Tract probability maps in stereotaxic spaces: analyses of white matter anatomy and tract-specific quantification”, Neuroimage,39:336–347:2008.
5. S. Bonavita, G. Tedeschi, A. Gallo, Morphostructural MRI abnormalities related to neuropsychiatric disorders associated to multiple sclerosis, Multiple Sclerosis International,2013.
6. P. Preziosa, E. Pagani, S. Mesaros et al., Progression of regional atrophy in the left hemisphere contributes to clinical and cognitive deterioration in multiple sclerosis: A 5-year study, Hum. Brain Mapp. 38(11):5648–5665:2017.
7. A. Ozturk, S.A. Smith, E.M. Gordon-Lipkin, et al., MRI of the Corpus Callosum in multiple sclerosis: association with disability, Multiple Sclerosis 16(2):166:2010.
8. Bloom, J.S., Hynd, G.W. The Role of the Corpus Callosum in Interhemispheric Transfer of Information: Excitation or Inhibition?. Neuropsychol Rev 15:59–71:2005.
Figures