0552
Motion-Resolved Brain MRI for Quantitative Multiparametric Mapping1Biomedical Imaging Research Institute, Cedars-Sinai Medical Center, Los Angeles, CA, United States
Synopsis
We introduce a motion-resolved solution to clinical brain MRI for quantitative multiparametric mapping using Multitasking. We demonstrate that the proposed approach is generalizable to translation, rotation, discrete motion, and periodic motion without explicit need for motion correction or compensation. Both simulation and in vivo results show that the proposed motion-resolved approach produces better image quality with sharp tissue structure and without ghosting/blurring artifacts, which outperforms no motion handling and simple motion removal. The motion-resolved approach yields substantially less RMSE in terms of quantitative mapping accuracy compared to no motion handling and simple motion removal.
Introduction
Motion poses a major challenge in clinical brain MRI. Nearly 30% of inpatient scans suffer from motion artifacts1. Numerous efforts have been made to address brain motion2-4, but no one solution has been generalizable enough to tackle all motion types5. Quantitative multiparametric mapping, which measures biomarkers offering complementary tissue information for disease evaluation6-7, is especially prone to motion artifacts due to prolonged scan times. Here, we introduce a stand-alone motion-resolved approach to quantitative multiparametric mapping using Multitasking, which is generalizable to translation, rotation, discrete motion, and periodic motion without explicit need for motion correction/compensation.Methods
Image model:Multitasking conceptualizes overlapping image dynamics as different time dimensions8, leading to an underlying image $$$x\left(\mathbf{r},t_{1},t_{2},\ldots,t_{N},s\right)$$$ with spatial index $$$\mathbf{r}$$$, motion state index $$$s$$$, and $$$N$$$ time index $$$t_{i}$$$ related to quantifiable parameters of interest (e.g., T1/T2/T1$$$\rho$$$/T2*/ADC). This image can be rearranged into an ($$$N$$$+2)-way low-rank9 tensor $$$\mathcal{X}$$$, which can be decomposed as10:
$$\mathcal{X}=\mathcal{V}\times_{1}\mathbf{U}_{r},$$
$$\mathcal{V}=\mathcal{C}\times_{2}\mathbf{U}_{t_{1}}\times_{3}\mathbf{U}_{t_{2}}\times_{4}\ldots\times_{N+1}\mathbf{U}_{t_{N}}\times_{N+2}\mathbf{U}_{s},$$
where $$$\mathcal{C}$$$ denotes the core tensor, each $$$\mathbf{U}$$$ is the factor matrix for the corresponding dimension, and $$$\times_{i}$$$ denotes $$$i$$$-mode tensor product11.
Image reconstruction:
Fig. 1 shows the image reconstruction pipeline.
First, an image with a single time dimension $$$t$$$ representing elapsed time, denoted as $$$\mathbf{X_{\mathrm{rt}}}$$$, is reconstructed using low-rank matrix strategy9,12.
Second, the last recovery time of each shot is extracted from $$$\mathbf{X_{\mathrm{rt}}}$$$, denoted as $$$\mathbf{X_{s}}$$$. Feature matrix $$$\mathbf{F}=\left(\mathbf{f}_{1},\mathbf{f}_{2},\ldots,\mathbf{f}_{N_{s}}\right)$$$ is extracted from $$$\mathbf{X_{s}}$$$ via PCA ($$$N_{s}$$$ denotes the number of recovery periods). To identify different motion states, k-means clustering is performed on $$$\mathbf{F}$$$. To select the number of motion states/clusters $$$K$$$, the algorithm is performed for $$$K$$$=1,2,…,20. Euclidean distance is calculated for each $$$K$$$:
$$d_{K}=\sum_{k=1}^{K}\sum_{\mathrm{f}_{s}\in{C_{k}}}\left\|\mathbf{f}_{s}-\mathbf{c}_{k}\right\|^{2},s\in\left\{1,2,\ldots,N_{s}\right\}$$
where $$$\mathbf{c}_{k}$$$ is the centroid of the $$$k$$$th cluster $$$C_{k}$$$. We choose $$$K$$$ from the elbow of $$$(K, d_{K})$$$ plot.
Third, the multidimensional factor $$$\hat{\mathcal{V}}$$$ is recovered from Bloch-constrained small-scale low-rank tensor completion of subspace training data $$$\mathbf{D_{\mathrm{tr}}}$$$8,12-13.
Fourth, to reduce the effect of outlier motion, a diagonal weighting matrix $$$\mathbf{W}$$$ is calculated from the residual comparing $$$\mathbf{D_{\mathrm{tr}}}$$$ to $$$\hat{\mathbf{V_{\mathrm{rt}}}}$$$ (i.e., $$$\hat{\mathcal{V}}$$$ mapped back to single-time domain):
$$\mathbf{R}=\mathbf{D}_{\mathrm{tr}}-\mathbf{D}_{\mathrm{tr}}\widehat{\mathbf{V}}_{\mathrm{rt}}^{\dagger}\widehat{\mathbf{V}}_{\mathrm{rt}},$$
$$W_{jj}=\left(\sum_{i}\left|R_{ij}\right|^{2}\right)^{-1/2}.$$
Fifth, $$$\mathbf{U}_{r}$$$ is estimated by solving:
$$\mathbf{U}_{r}=\arg\min_{\mathbf{U}_{r}}\left\|\mathbf{W}\left[\mathbf{d}_{\mathrm{img}}-E\left(\hat{\mathcal{V}}\times_{1}\mathbf{U}_{r}\right)\right]\right\|^{2}+R_{s}\left(\mathbf{U}_{r}\right),$$
where $$$\mathbf{d}_{\mathrm{img}}$$$ represents the vectorized imaging data8,12-13, $$$E$$$ combines multichannel encoding and sampling, and $$$R_{s}(\cdot)$$$ performs spatial regularization.
Lastly, multiparametric fitting8,12-13 is performed on $$$\mathcal{X}$$$ selecting the most-populous motion state.
Motion simulation:
3D whole-brain T1/T2/T1$$$\rho$$$ maps13 were used to generate a numerical motion phantom which was used to simulate 15min Multitasking scan13. Two motion schemes were investigated including 8 scenarios:
Discrete motion: 6 scenarios were simulated with 2,3,4,5,6,12 discrete motion states. For each scenario, three translations and three rotations along and around xyz axis were selected from [-8mm,8mm] and [-8°,8°] following Gaussian distribution for each motion state.
Periodic motion: 2 scenarios were simulated where z-rotation from [-8°,8°] (simulating head shaking) were periodically added during the middle 5min of the simulated scan. For one scenario, the head returned to the original 0° position after motion; for the other, the head rotated to a different 5° position after motion.
For each scenario, we went through the estimation of $$$K$$$ and motion state clustering.
In vivo experiments:
Data were collected on $$$n$$$=2 healthy subjects on Siemens Biograph mMR scanner. A Multitasking T1/T2/T1$$$\rho$$$ mapping sequence13 was implemented with FOV=240x240x140mm3, resolution=1.0x1.0x3.5mm3, scan time=7min. For each subject, one motion-free scan was performed followed by two motion scans: one containing discrete motion for at least 2 movements, the other containing periodic motion for at least 2min. Each subject was given instructions of the motion type and the minimum amount of motion to be performed.
Evaluations:
Three reconstructions were compared: i) no motion-handling, where all k-space data were used for reconstruction assuming a single motion state ($$$K$$$=1); ii) motion-removal, where k-space data outside of the most-populous state were discarded; iii) motion-resolved, as described above. For the simulations, RMSE of reconstructed T1/T2/T1$$$\rho$$$ maps compared to phantom ground truth were calculated for evaluation.
Results
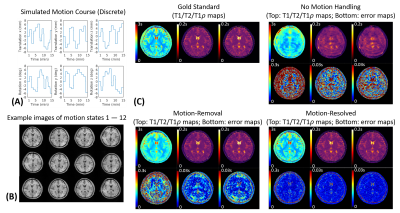
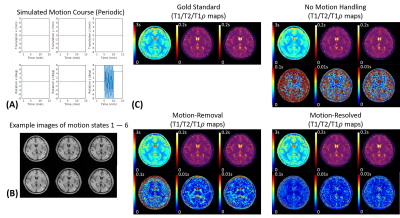
Fig.2A shows the simulated motion pattern for the discrete motion scenario with 12 states. Our algorithm correctly determined $$$K$$$=12, and all those states were successfully identified (Fig.2B). The motion-resolved solution produced substantially reduced absolute difference errors compared to no motion-handling and motion-removal (Fig.2C).Fig.3A shows the motion pattern for the periodic motion scenario with 5 ending position. $$$K$$$=6 motion states were binned by our algorithm (Fig.3B). Again, the motion-resolved solution produced substantially reduced absolute difference errors against the gold standards (Fig.3C).
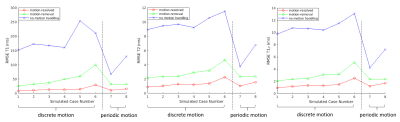
Fig.4 shows the RMSE of the reconstructed T1/T2/T1$$$\rho$$$ maps against the gold standards for all 8 simulated scenarios. The proposed solution substantially reduced RMSE for T1/T2/T1$$$\rho$$$.
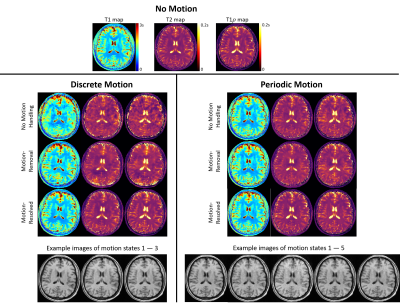
Fig.5 shows example in vivo results for discrete and periodic motion. Three motion states were identified for discrete motion, and 5 motion states were identified for periodic motion. For both experiments, the motion-resolved solution produced T1/T2/T1$$$\rho$$$ maps with the best image quality with less ghosting/blurring artifacts and sharper tissue structures.
Discussion and Conclusion
We introduced a motion-resolved solution for quantitative brain multiparametric mapping, which outperformed no motion-handling and motion-removal. Feasibility was demonstrated for T1/T2/T1$$$\rho$$$ mapping, with extension to other quantifiable tissue parameters straightforward using Multitasking. The proposed solution works as a stand-alone approach which is generalizable to translation, rotation, discrete motion, and periodic motion, with no explicit need for motion correction/compensation.Acknowledgements
This work was supported by NIH 1R01EB028146.References
1. Andre JB, Bresnahan BW, Mossa-Basha M, Hoff MN, Smith CP, Anzai Y, et al. Toward Quantifying the Prevalence, Severity, and Cost Associated With Patient Motion During Clinical MR Examinations. J Am Coll Radiol. 2015;12(7):689-695.
2. Godenschweger F, Kagebein U, Stucht D, Yarach U, Sciarra A, Yakupov R, et al. Motion correction in MRI of the brain. Phys Med Biol. 2016;61(5):R32-56.
3. Thesen S, Heid O, Mueller E, Schad LR. Prospective acquisition correction for head motion with image-based tracking for real-time fMRI. Magn Reson Med. 2000;44(3):457-465.
4. Maclaren J, Herbst M, Speck O, Zaitsev M. Prospective motion correction in brain imaging: a review. Magn Reson Med. 2013;69(3):621-636.
5. Zaitsev M, Maclaren J, Herbst M. Motion artifacts in MRI: A complex problem with many partial solutions. J Magn Reson Imaging. 2015;42(4):887-901.
6. Badve C, Yu A, Dastmalchian S, et al. MR Fingerprinting of Adult Brain Tumors: Initial Experience. AJNR Am J Neuroradiol. 2017;38(3):492-499.
7. Hattingen E, Jurcoane A, Daneshvar K, et al. Quantitative T2 mapping of recurrent glioblastoma under bevacizumab improves monitoring for non-enhancing tumor progression and predicts overall survival. Neuro Oncol. 2013;15(10):1395-1404.
8. Christodoulou AG, Shaw JL, Nguyen C, et al. Magnetic resonance multitasking for motion-resolved quantitative cardiovascular imaging. Nat Biomed Eng. 2018;2(4):215-226.
9. Liang Z-P. Spatiotemporal imagingwith partially separable functions. In Proceedings of the 4th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Arlington, Virginia, USA, 2007. p. 988-991.
10. Tucker LR. Some mathematical notes on three-mode factor analysis. Psychometrika. 1966;31(3):279-311.
11. He J, Liu Q, Christodoulou A, Ma C, Lam F, Liang Z-P. Accelerated high-dimensional MR imaging with sparse sampling using low-rank tensors. IEEE Trans Med Imaging. 2016;35(9):2119-2129.
12. Ma S, Nguyen CT, Han F, Wang N, Deng Z, Binesh N, et al. Three-dimensional simultaneous brain T1 , T2 , and ADC mapping with MR Multitasking. Magn Reson Med. 2020;84(1):72-88.
13. Ma S, Wang N, Fan Z, Kaisey M, Sicotte NL, Christodoulou AG, et al. Three-Dimensional Whole-Brain Simultaneous T1, T2, and T1 Quantification using MR Multitasking: Method and Initial Clinical Experience in Tissue Characterization of Multiple Sclerosis. Magn Reson Med. 2020; In Press.
Figures