0531
Automatic segmentation of uterine endometrial cancer on MRI with convolutional neural network1Diagnostic Imaging and Nuclear Medicine, Kyoto university hospital, Kyoto, Japan, 2Preemptive Medicine and Lifestyle-Related Disease Research Center, Kyoto university hospital, Kyoto, Japan, 3Real World Data Research and Development, Graduate School of Medicine Kyoto University, Kyoto, Japan, 4Diagnostic Pathology, Kyoto university hospital, Kyoto, Japan, 5Gynecology and Obstetrics, Kyoto university hospital, Kyoto, Japan
Synopsis
Endometrial cancer is the most common gynecological malignant tumor in developed countries, and accurate preoperative risk stratification is essential for personalized medicine. For realizing tumor feature extraction by radiomics approach, the segmentation of the tumor is usually required. The model developed in this study has achieved high-accuracy automatic segmentation of endometrial cancer on MRI using a convolutional neural network for the first time. Using multi-sequence MR images were important for high accuracy segmentation. Our model will lead to efficient medical image analysis of a large number of cases using the radiomics approach and/or deep learning methods.
INTRODUCTION
Endometrial cancer is the most common gynecological malignant tumor in developed countries and its incidence rates have been increasing over the past decades1. The treatment strategy for endometrial cancer depends on the tumor stage and patients’ risk of recurrence. Accurate methods of preoperative risk stratification are essential to prevent both overtreatment and undertreatment, which leads to personalized medicine. There are also some reports where the radiomics approach is applied to endometrial cancer to stratify the risk of patients2-6. For performing these researches, tumor segmentation on MRI is usually necessary. As manual segmentation is not only labor-intensive, time-consuming, but sometimes subjective, accurate automated segmentation methods are highly desirable in medical image analysis with a large dataset.U-net is a fully convolutional neural network architecture originally designed for biomedical image segmentation, which has shown promising results in medical image segmentation7. In the field of obstetrics and gynecology, some reports where U-net was applied for the segmentation of uterus and uterine cervical cancer, and both reported good segmentation performance8,9. There is no previous report on automatic segmentation of endometrial cancer using a convolutional neural network on MRI.
The purpose of this research was to realize the automatic segmentation of endometrial cancer on MRI with U-net modified for endometrial cancer. We also examined whether to use a single-sequence or multi-sequence images as the input data of U-net for higher segmentation accuracy.
METHODS
From 200 patients diagnosed with endometrial cancer pathologically, 180 patients were randomly selected as the training dataset, and the other 20 patients as the test dataset to evaluate the performance of the final model. MR images used in this study were sagittal T2-weighted image (T2WI), diffusion-weighted image (DWI), and apparent diffusion coefficient (ADC) map obtained at 1.5T or 3T machines. To evaluate the effect of using multi-sequence of MR images for improving segmentation accuracy, single-sequence images (one of T2WI, DWI, and ADC map), and multi-sequence images (T2WI-DWI-ADC map for triple channel) were used as input data. ROIs of endometrial cancer segmented by two radiologists by consensus were the reference standard. Our modified U-net architecture for the segmentation of endometrial cancer is presented in Figure 1. Dice loss function was used as the cost function. Five-fold cross-validation was performed for training the model parameters. For comparison, a conventional U-net model was trained for the segmentation. Our model was built with Keras (ver. 2.3.1) and Tensorflow (ver. 1.15.0), and the model was trained on a personal computer with two NVIDIA Quadro RTX8000 GPU (NVIDIA, Santa Clara, CA, USA).The segmentation accuracy for the test datasets was evaluated using the Dice similarity coefficient (DSC), sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) for each patient. DSC of the test datasets with the different MRI sequences as input data was compared using Wilcoxon signed-rank test. A p-value < 0.05 was considered to indicate statistical significance. All p-values were adjusted for multiple comparisons by using the Holm method10.
RESULTS
The Dice loss of our models for five-fold cross-validation with input data of each MRI sequence is presented in Figure 2. The model with multi-sequence images as input data achieved the lowest Dice loss for both training and validation sets. The mean and standard deviation of Dice loss of the conventional U-net model with multi-sequence images as input data were 0.267±0.027 for five-fold cross-validation sets, which was worse than our modified U-net. The segmentation accuracy of our model for the test datasets with each input data is shown in Figure 3. The model with multi-sequence images achieved the highest DSC, Sensitivity, and PPV than the other models (DSC, sensitivity, and PPV were 0.806± 0.155, 0.816 ± 0.212, 0.834±0.086, respectively). There was no statistically significant difference between the model with multi-sequence images and the model with T2WI (p=0.67), but there was a statistically significant difference between the model with multi-sequence images and the model with DWI and the model with ADC map (p<0.001). Representative results of automatic segmentation are presented in Figures 4 and 5.DISCUSSION & CONCLUSION
We have achieved high-accuracy automatic segmentation of endometrial cancer on MRI using the modified U-net. For input data of the U-net, high accuracy was achieved by using multi-sequence images including T2WI, DWI, and ADC map. Although endometrial cancer usually shows high signal intensity on DWI due to its high cellularity, normal uterine endometrium also shows high signal intensity on DWI11. Then, radiologists recognize the extent of endometrial cancer by referring to various sequence images in clinical practice. This could be the reason why the model with multi-sequence images showed higher accuracy than the other models.Our model for the automatic segmentation of endometrial cancer would make it possible to prepare a large number of regions of interest (ROIs) for endometrial cancer with less effort, which leads to efficient medical image analysis using the radiomics approach and/or deep learning methods for the risk stratification of endometrial cancer.
Acknowledgements
This work was supported by JSPS KAKENHI Grant Number JP20K16780References
1. Morice P, Leary A, Creutzberg C, Abu-Rustum N, Darai E. Endometrial cancer. Lancet (London, England) 2016;387:1094-108.
2. Yan BC, Li Y, Ma FH, et al. Preoperative Assessment for High-Risk Endometrial Cancer by Developing an MRI- and Clinical-Based Radiomics Nomogram: A Multicenter Study. Journal of magnetic resonance imaging : JMRI 2020;52:1872-82.
3. Fasmer KE, Hodneland E, Dybvik JA, et al. Whole-Volume Tumor MRI Radiomics for Prognostic Modeling in Endometrial Cancer. Journal of magnetic resonance imaging : JMRI 2020.
4. Stanzione A, Cuocolo R, Del Grosso R, et al. Deep Myometrial Infiltration of Endometrial Cancer on MRI: A Radiomics-Powered Machine Learning Pilot Study. Academic radiology 2020.
5. Ytre-Hauge S, Dybvik JA, Lundervold A, et al. Preoperative tumor texture analysis on MRI predicts high-risk disease and reduced survival in endometrial cancer. Journal of magnetic resonance imaging : JMRI 2018;48:1637-47.
6. Ueno Y, Forghani B, Forghani R, et al. Endometrial Carcinoma: MR Imaging-based Texture Model for Preoperative Risk Stratification-A Preliminary Analysis. Radiology 2017;284:748-57.
7. Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. arXiv e-prints. arXiv preprint arXiv:150504597 2015.
8. Kurata Y, Nishio M, Kido A, et al. Automatic segmentation of the uterus on MRI using a convolutional neural network. Computers in biology and medicine 2019;114:103438.
9. Lin YC, Lin CH, Lu HY, et al. Deep learning for fully automated tumor segmentation and extraction of magnetic resonance radiomics features in cervical cancer. European radiology 2020;30:1297-305.
10. Holm S. A Simple Sequentially Rejective Multiple Test Procedure. Scandinavian Journal of Statistics 1979;6:65-70.
11. Tamai K, Koyama T, Saga T, et al. Diffusion-weighted MR imaging of uterine endometrial cancer. Journal of magnetic resonance imaging : JMRI 2007;26:682-7.
Figures
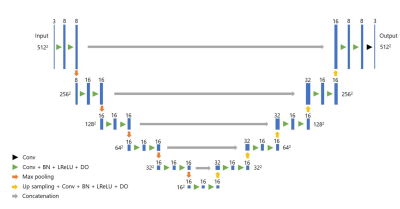
Figure 1: Our U-net architecture
Conv: convolution
BN: Batch normalization
LReLU: Leaky Rectified Linear Unit
DO: Dropout
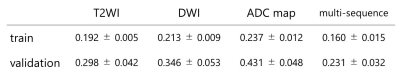
Figure 2: Dice loss of the models with each MRI sequence as input data for five-fold cross-validation
Data are presented with mean±standard deviation of five cross-validation models.
T2WI: T2-weighted image
DWI: diffusion-weighted image
ADC: apparent diffusion coefficient
Multi-sequence: T2WI, DWI, and ADC map
train: train loss
validation: validation loss
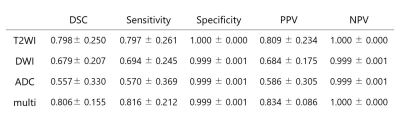
Figure 3: The segmentation accuracy of our model for the test datasets with each MRI sequences as input data
Data are presented with mean±standard deviation
T2WI: T2-weighted image
DWI: diffusion-weighted image
ADC: apparent diffusion coefficient
Multi: T2WI, DWI, and ADC map
DSC: Dice similarity coefficient
PPV: positive predictive value
NPV: negative predictive value
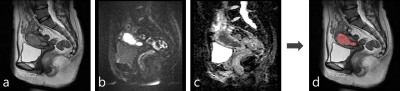
Figure 4: A representative case of the automatic segmentation
a: T2-weighted image
b: diffusion-weighted image (b=1000 s/mm2)
c: apparent diffusion coefficient map
d: A result of automatic segmentation of endometrial cancer overlaid on T2-weighted image
The tumor was almost perfectly segmented (Dice similarity coefficient=0.925).

Figure 5: A representative case of the automatic segmentation
a: T2-weighted image
b: diffusion-weighted image (b=1000 s/mm2)
c: apparent diffusion coefficient map
d: A result of automatic segmentation of endometrial cancer overlaid on T2-weighted image
The tumor was well segmented despite the presence of hematometra (a:*) (Dice similarity coefficient=0.808).