0466
LayNii: A software suite for layer-fMRI1Faculty of Psychology and Neuroscience, Maastricht University, Maastricht, Netherlands, 2NIH, Bethesda, MD, United States, 3UCL, London, United Kingdom, 4CMRR, Minneapolis, MN, United States, 5University of Glasgow, Glasgow, United Kingdom, 6Uni Mainz, Mainz, Germany, 7Scannexus, Maastricht, Netherlands, 8Brain Innovation, Maastricht, Netherlands
Synopsis
- A new software toolbox is introduced for layer-specific functional MRI: LayNii.
- LayNii is a suite of command-line executable C++ programs. It can be easily installed via source code and binaries for Linux, Windows, and macOS.
- LayNii is designed for layer-fMRI data that suffer from SNR and coverage constraints and facilitates user-dependent parameter tweaking.
- LayNii performs layerification, columnar distance estimation, and cortical unfolding in the native voxel space of functional data.
- LayNii performs layer-smoothing, GE-BOLD deveining, fMRI quality assessment, and VASO analysis.
Purpose
High-resolution fMRI in the sub-millimeter regime allows researchers to resolve brain activity across cortical layers and columns non-invasively (Koopmanns 2010, Olman 2012, Huber 2017). While these high-resolution data make it possible to address novel questions of directional information flow within and across brain circuits, the corresponding data analyses are challenged by MRI acquisition constraints including: low SNR, image distortions, and restricted coverage (Weldon 2020). These challenges often result in insufficient performance accuracy of conventional analysis pipelines and can only partly be mitigated with time consuming manual interventions and additional data acquisition of various auxiliary analysis facilitating images. Here we introduce a new software suite specifically designed for layer-specific functional MRI and all its current limitations: LayNii.The purpose of LayNii:
- Performing layer-fMRI analyses directly in native EPI space without being limited by suboptimal registration or distortion-correction performance at the sub-voxel accuracy level.
- Performing laminar and columnar topography analyses fully in voxel space without the need for surface approximations. Working in voxel space circumvents conventional requirements of topographical requirements of continuous, closed surfaces that are rarely met with small FOV layer-fMRI protocols.
- Performing spatial smoothing without losing precious information of the fine scale layer-fMRI activity across cortical depth.
Methods
This non-commercial toolbox is a collection of command-line executable programs written in C++. It is distributed as open-source code and as pre-compiled binaries for Linux, Windows, and macOS (without external dependencies). LayNii is designed for layer-fMRI data that suffer from SNR and coverage constraints and thus cannot be straightforwardly analysed in alternative software packages. Some of the most popular programs of LayNii contain ‘layerification’ and columnarization in the native voxel space of functional data as well as many other layer-fMRI specific analysis tasks: layer-specific smoothing, model-based mitigation of vein effects in GE-BOLD data, quality assessment of artifact-dominated sub-millimeter fMRI, as well as analyses of VASO data. Since LayNii works directly in voxel space without imposing topological constraints, it can straightforwardly work in the native EPI space of functional fMRI. It does not require closed surfaces of entire hemispheres and works straightforwardly in isotropic, as well as non-isotropic voxels, and even on 2D-data.LayNii’s functionalities
Figures 1-5 exemplify the features of LayNii's most useful functionalities:- Fig. 1: Assigning layer values to each voxel across the cortical depth. aka. Layerification.
- Fig. 2: Column assignment for topographical layer analyses.
- Fig. 3: Layer specific smoothing similar to (Blazejewska 2019).
- Fig. 4: Quality-Assessment of sub-millimeter fMRI data.
- Fig. 5: Model-based mitigation of draining vein effects.
Discussion
The current functionality largely focuses on applications where the challenges of layer-fMRI data do not allow the application of standard analysis pipelines of the major software packages. It does not duplicate some analysis features that are already in other fMRI software packages (e.g., registration and segmentation in: AFNI, Brain Voyager, SPM, FreeSurfer, Nighres, CBS tools etc.). The modular structure of LayNii allows for easy integration with analysis pipelines for other software packages.LayNii is designed to provide mechanisms, not policies. It is kept as modular as possible giving the user increased flexibility for manual interventions and algorithm fine tunings. However, this package structure also means that LayNii does not give "one-size-fits-all" solutions but requires the user to understand what its programs with all its tuning parameters do.
Acknowledgements
We thank Pilou Bazin for contributing comments, discussions and ideas regarding the algorithms and implementation of the equi-volume layerification and layer-specific smoothing programs of LayNii. We thank Kamil Uludag and Martin Havlicek for comments on the manuscript regarding the model-based deveining.
Funding: Parts of this research was supported by the NIMH Intramural Research Program (ZIA-MH002783). Laurentius Huber was funded form the NWO VENI project 016.Veni.198.032 for part of the study. Benedikt Poser is partially funded by the NWO VIDI grant 16.Vidi.178.052 and by the National Institute for Health grant (R01MH/111444) (PI David Feinberg). Portions of this study used the high- performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (biowulf.nih.gov). Rainer Goebel is partly funded by the European Research Council Grant ERC-2010-AdG 269853 and Human Brain Project Grant FP7-ICT-2013-FET-F/604102. Nils Nothnagel and Jozien Goense are funded by the Medical Research Council (MR/R005745/1). Andrew T. Morgan is funded by the Medical Research Council (MR/N008537/1) and the European Union’s Horizon 2020 Framework Programme for Research and Innovation under the Specific Grant Agreement No. 785907 and 945539 (Human Brain Project SGA2 and SGA3).
Discussions of code: We thank Sriranga Kashyap for many informative questions of previous versions of LAYNII. We thank Kamil Uludag for helpful discussions and contributions on model-based-vein deveining. We thank Elisha Merriam for discussions about the least noise amplifications in LN_ BOCO with spline surround-division. We thank the many users of LAYNII that submitted bug reports and feature requests to our repository.
Acquisition of example data: We thank Kenny Chung and Joe Stolinski for radiographic assistance with experiments conducted at NIH. We thank Sean Marrett for co-acquiring data used in Fig. 2-5. We thank FMRIF (especially Andy Derbyshire), and Scannexus (especially Chris Wiggins), where example data were acquired.
Financial interest: Authors OFG and RG work for Brain Innovation and have financial interest tied to the company.
Betha users: We thank the many LAYNII users who sent feedback via Github issues. We also thank Dimo Ivanov, Sri Kashyap and Deni Kurban for testing the binaries.
References
- Amunts K, Lepage C, Borgeat L, et al. BigBrain: An ultrahigh-resolution 3D human brain model. Science (80- ). 2013;340(6139):1472-1475. doi:10.1126/science.1235381Blazejewska, Neuroimage. 2019; doi:10.1016/j.neuroimage.2019.01.054
- Ding SL, Royall JJ, Sunkin SM, et al. Comprehensive cellular-resolution atlas of the adult human brain. J Comp Neurol. 2016;524:3127-3481. doi:10.1002/cne.24080Havlicek M, Uludag K. A dynamical model of the laminar BOLD response. Neuroimage. 2019;204:116209. doi:10.1101/609099
- Heinzle J, Koopmans PJ, den Ouden HEM, Raman S, Stephan KE. A hemodynamic model for layered BOLD signals. Neuroimage. 2016;125:556-570. doi:10.1016/j.neuroimage.2015.10.025
- Huang P, Correia MM, Rua C, Rodgers CT, Henson N, Carlin JD. Correcting for Superficial Bias in 7T Gradient Echo fMRI. bioRxiv. 2020. doi:https://doi.org/10.1101/2020.11.20.392258
- Huber L, Handwerker DA, Jangraw DC, et al. High-Resolution CBV-fMRI Allows Mapping of Laminar Activity and Connectivity of Cortical Input and Output in Human M1. Neuron. 2017;96(6):1253-1263.e7. doi:10.1016/j.neuron.2017.11.005
- Koopmans PJ, Barth M, Norris DG. Layer-specific BOLD activation in human V1. Hum Brain Mapp. 2010;31(9):1297-1304. doi:10.1002/hbm.20936
- Lacy TC, Robinson PA, Aquino KM, Pang JC. Cortical Depth-Dependent Modeling of Visual Hemodynamic Responses. bioRxiv. 2020. doi:10.1101/2020.03.16.993154,
- Markuerkiaga I, Marques JP, Gallagher TE, Norris DG. Estimation of Laminar BOLD Activation Profiles using Deconvolution with a Physiological Point Spread Function. bioRxiv. 2020:1-28. doi:10.1101/2020.08.04.236190
- Markuerkiaga I, Barth M, Norris DG. A cortical vascular model for examining the specificity of the laminar BOLD signal. Neuroimage. 2016;132:491-498. doi:10.1016/j.neuroimage.2016.02.073
- Olman CA, Harel N, Feinberg DA, et al. Layer-Specific fMRI Reflects Different Neuronal Computations at Different Depths in Human V1. PLoS One. 2012;7(3):e32536. doi:10.1371/journal.pone.0032536
- Wagstyl K, Larocque S, Cucurull G, et al. Automated segmentation of cortical layers in BigBrain reveals divergent cortical and laminar thickness gradients in sensory and motor cortices. PLOS Biol. 2020;18(4):e3000678. doi:10.1101/580597
- Weldon KB, Olman CA, Olman CA. Forging a path to mesoscopic imaging success with ultra-high field functional magnetic resonance imaging. Philos Trans B. 2020.
Figures
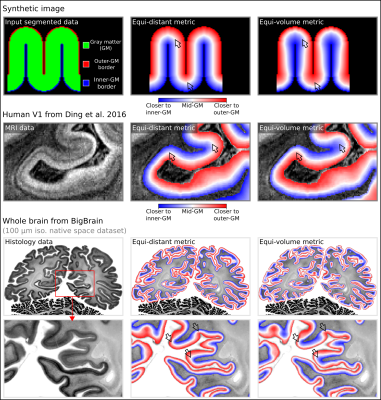
Layering metrics generated in LayNii:
The top row shows an application with a synthetic 2D image. The middle row shows the empirical layers from Ding et al. (2016) (0.2 mm iso.). The bottom row shows BigBrain (0.1 mm iso., native space) (Amunts et al. 2013) with cortical borders provided in Wagstyl et al. (2020). The equi-distant metric is shown in the middle column and equi-volume metric is shown in the right column for each image type. The arrows highlight areas where the equi-distant and the equi-volume metric differ considerably.
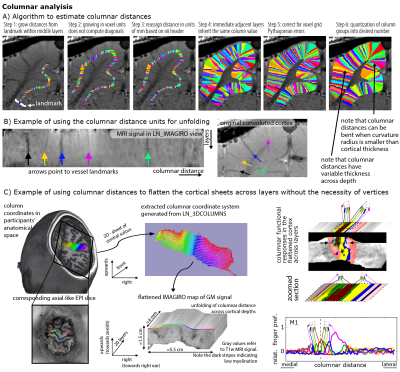
Estimating columnar units in voxel space with LayNii:
Panel A) describes the algorithm of estimating columnar distances in the example of a cat visual cortex.
Panel B) depicts cortical unfolding in LayNii. The orthogonal coordinates of columnar distances and cortical depths are used to map the signal intensity on a new spatial grid and then LayNii writes it out as NIFTI files.
Panel C) depicts an application for topographic mapping in the human somatosensory system of depicting digit representation in the anterior bank of the central sulcus acros laminar and columnar directions.
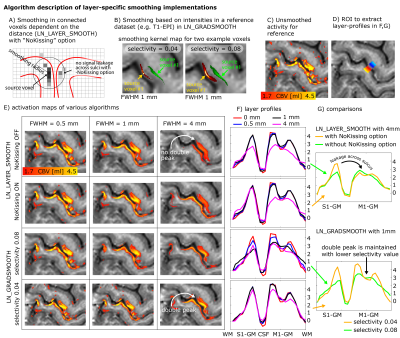
Panels A-B) describe two different algorithms of layer-smoothing. A) depicts danger of signal leakage across kissing gyri. B) depicts example smoothing kernels in contrast-specific smoothing. The smoothing kernels are wider in voxels of similar anatomical contrast.
E-G) show different results of the respective smoothing methods. Namely smoothing within layers that are parallel to GM/WM and GM/CSF border lines (top rows) and smoothing based on similar MRI signal intensities (bottom rows).
A characteristic double peak pattern is used as a test bed to assess the signal blurring.
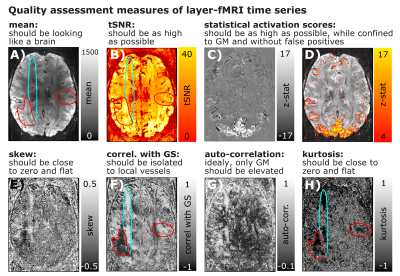
Submillimeter fMRI QA:
Panels A-D) depict maps of conventional fMRI QA metrics of Mean, tSNR, and activation scores.
Panels E-H) depict higher-order QA metrics that can be informative in layer-fMRI. While the QA measures in the top row suggest that the underlying time series is of high quality, higher-order QA measures in the bottom row show that there are indeed typical phase errors and MR-artifacts present (turquoise and red).
While these artifacts are clearly visible in most quality metrics of E-H), they are hardly visible in first order quality metrics A-D).
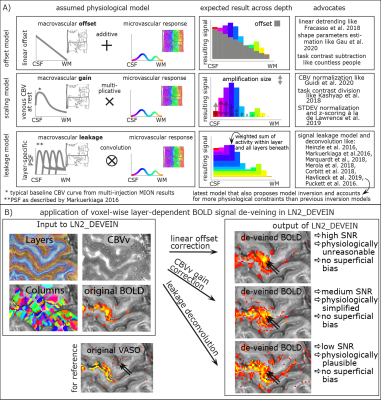
Model-based mitigation of draining vein effects:
While model-based vein removal methods are still being validated and suffer from countless simplifications as well as an abundance of free parameters, their application for exploratory use is available in LayNii in three possible ways. All of them mitigate the superficial vein bias. However, they come along with variable noise amplification, and partly also introduce unwanted autocorrelation of noise.