0453
Voxel-wise Tracking of Grid Tagged Cardiac Images using a Neural Network Trained with Synthetic Data1Radiology, Stanford University, Stanford, CA, United States, 2Radiology, Veterans Affairs Health Care System, Palo Alto, CA, United States, 3Mechanical and Aerospace Engineering, University of Central Florida, Orlando, FL, United States, 4Maternal & Child Health Research Institute, Stanford University, Stanford, CA, United States, 5Cardiovascular Institute, Stanford University, Stanford, CA, United States
Synopsis
This work introduces a neural network for tracking myocardial motion in cine grid tagged MR images on a voxel-wise basis. This is achieved with the use a synthetic training dataset that includes comprehensive motion patterns. Synthetic training allows for a known ground truth motion to be included in training. The network was tested against a previous network that tracked only tag line intersections. Displacements and strain maps were generated and compared. The voxel tracking network shows qualitatively better spatial localization of strain, and better radial strain values compared to tracking only tag lines.
Introduction
Cine grid tagged cardiac MRI enables the measurement of cardiac displacements and the quantification of cardiac strains, which are an import biomarker of cardiac function and dysfunction. By tracking the tag lines placed on the image, displacements can be measured. However, tracking tag lines can be cumbersome, requiring manual review and intervention. Machine learning has shown to be a promising technique for motion tracking applied to this problem.1,2Previously, a comprehensive synthetic data generator for training a neural network to accurately track tag line intersections and compute strain has been demonstrated.2 Using synthetic data is promising as it contains a known ground truth, is easy to generate and modify, and is not constrained by regulatory considerations nor biased by inclusion demographics. In this work, we expand upon this methodology to demonstrate that synthetic data training can also enable voxel-wise tracking of the myocardium. This potentially allows for greater spatial localization of displacements, and therefore strain. Our objective was to demonstrate the use of a neural net based, voxel-wise tracking technique for the effective quantification of local strains from grid tagged images.
Methods
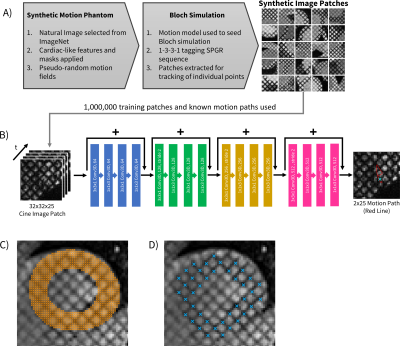
A synthetic data generation algorithm was used to generate time-resolved deforming natural images for training with known displacement values for all points and timeframes. This data generator then uses a Bloch simulation to produce a cine grid tagged dataset with realistic image quality (Fig 1A). The images and their respective motion paths were used to train an 18-layer Resnet architecture3 with modifications to include both coordinate convolutions4 and (2+1)D convolutions5, therefore enabling convolutions in both the spatial and temporal dimensions. The network tracks a single point from a surrounding 32x32 voxel patch. Each individual point to be tracked is processed by the network to extract its specific motion path (Fig 1B). 10^6 training patches were used to train the network, and inference takes ~1 second to track all voxels in a time-resolved series.The methods were applied in short-axis, mid-LV slices in healthy pediatric subjects (N=9) with IRB approval and consent. Cine grid tagged images were acquired with: TE/TR=2.5ms/4.9ms, flip angle=10°, FOV=260mmx320mm, 110° total tagging flip angle, spatial resolution 1.4mmx1.4mmx8mm, 25 time frames, 8-12s breath hold. The LV was manually segmented in the first time frame and all voxels in the mask were used for subsequent automated tracking (Fig 1C). Additionally, the tag line intersections were manually delineated and tracked with a separate network trained only on intersection tracking for comparison (Fig 1D). This approach has been well validated for accurately tracking tag line intersections, as well as computing strain.2
Tracked points were visualized with displacement vectors, as well as displacement maps, which were generated by linearly interpolating the tracked points onto an imaging grid. Strains were calculated by differentiating the displacement field interpolated using a radial basis function with a Gaussian kernel and shape parameter = 1.4mm (1 pixel) for voxel tracking and 8mm (equivalent to tag spacing) for tag line intersection tracked images.6 Circumferential (Ecc) and radial (Err) strains were investigated as maps, as well as the mean global strain values across the LV ROI.
Results
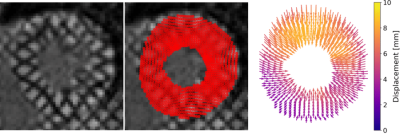
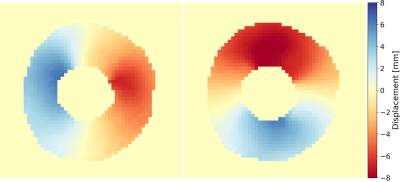
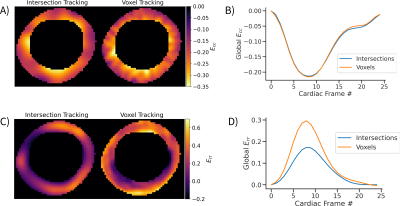
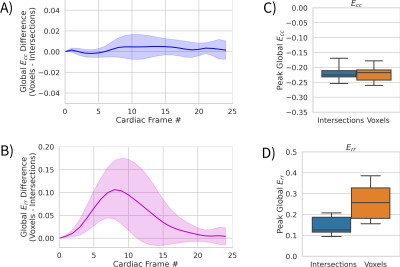
Figure 2 shows an animation of the tracking output from the neural network, shows tracked points, and the corresponding displacement vectors relative to the reference configuration (t=0). Figure 3 shows an animation of the displacement values interpolated onto an image grid. When the tracking the same locations (tag intersections), the two networks differed by RMSE = 0.29mm. Figure 4 shows an example case comparing intersection tracking to voxel-wise tracking. Fig. 4A shows Ecc maps, where similar patterns and values are seen, with qualitatively less blurring in the voxel tracked maps. Fig. 4B shows a global Ecc curve from the same case, where similar values are seen. Fig. 4C shows Err images from both techniques, and Fig 4D shows the corresponding global Err curves from both methods. In this example, Err is higher with the voxel-wise tracking as compared to intersection tracking. Figure 5A and 5B show strain differences across all subjects. Fig. 5C and 5D show box plots of peak global Ecc and Err for all subjects. In both instances good agreement is seen for Ecc, and voxel-wise tracking reports higher Err values.Discussion
In this work, we use synthetically generated data to train a neural network for tracking every voxel in the myocardium of grid-tagged images. The use of synthetic data enables the direct and quantitative comparison of alternative strategies (tracking voxels versus intersections) since the ground truth motion is known at all points in the training data, which is otherwise difficult or impossible to obtain with real data. Compared to tracking tag line intersections, voxel-wise tracking allows for better spatial localization of the displacement and strain values. This is seen qualitatively in the strain maps and in the radial strain values. Tracking intersections results in reduced Err, most likely because there are not enough points in the radial direction to estimate it properly. Notably, the use of voxel-wise tracking gives more reasonable Err values for healthy subjects (20%-40%).7 Further work is needed to compare to other tracking methods, as well as in a larger patient cohort with cardiac dysfunction.Acknowledgements
R01 HL131823, R01 HL131975, R01 HL152256References
[1] Ferdian, Edward, et al. "Fully Automated Myocardial Strain Estimation from Cardiovascular MRI–tagged Images Using a Deep Learning Framework in the UK Biobank." Radiology: Cardiothoracic Imaging 2.1 (2020): e190032.
[2] Loecher M, Perotti LE , Ennis DB. Cardiac Tag Tracking with Deep Learning Trained with Comprehensive Synthetic Data Generation. ISMRM 28th Annual Meeting, Paris, France (Virtual Conference), 2020
[3] He, Kaiming, et al. "Deep residual learning for image recognition." Proceedings of the IEEE conference on computer vision and pattern recognition. 2016.
[4] Liu, Rosanne, et al. "An intriguing failing of convolutional neural networks and the coordconv solution." Advances in neural information processing systems 31 (2018): 9605-9616.
[5] Tran, Du, et al. "A closer look at spatiotemporal convolutions for action recognition." Proceedings of the IEEE conference on Computer Vision and Pattern Recognition. 2018.
[6] Bistoquet, Arnaud, John Oshinski, and Oskar Škrinjar. "Myocardial deformation recovery from cine MRI using a nearly incompressible biventricular model." Medical image analysis 12.1 (2008): 69-85.
[7] Scatteia, A., A. Baritussio, and C. Bucciarelli-Ducci. "Strain imaging using cardiac magnetic resonance." Heart failure reviews 22.4 (2017): 465-476.
Figures