0449
MyoMapNet: A Deep Neural Network for Accelerating the Modified Look-Locker Inversion Recovery Myocardial T1 Mapping to 5 Heart Beats
Hossam El-Rewaidy1,2, Rui Guo1, and Reza Nezafat1
1Medicine, Beth Israel Deaconess Medical Center and Harvard Medical School, Boston, MA, United States, 2Graduate School of Bioengineering, Department of Computer Science, Technical University of Munich, Munich, Germany
1Medicine, Beth Israel Deaconess Medical Center and Harvard Medical School, Boston, MA, United States, 2Graduate School of Bioengineering, Department of Computer Science, Technical University of Munich, Munich, Germany
Synopsis
In this work, we developed and evaluated a rapid (4-5 heartbeats) myocardial T1 mapping approach by estimating voxel-wise T1 values from one look-locker (LL) experiment of MOLLI sequence using a fully-connected neural network (MyoMapNet). MyoMapNet consists of 5 hidden layers that map the input 4-5 T1-weighted samplings and their inversion times into T1 values. MyoMapNet was trained and evaluated on a large dataset of native MOLLI-5(3)3 T1 in 717 subjects and post-contrast MOLLI-4(1)3(1)2 in 535 subjects. MyoMapNet showed similar T1 estimations to MOLLI-5(3)3 and MOLLI-4(1)3(1)2 T1 (mean difference=1±17ms, and -3±18ms, respectively, p-value >0.1 for both).
Introduction
Cardiac T1 mapping using the Modified Look-Locker inversion recovery (MOLLI) allows non-invasive quantification of interstitial diffuse fibrosis (1). The original MOLLI-3(3)3(3)5 scheme consists of three LL experiments with a waiting period of 3 rest heartbeats between each LL for magnetization recovery (2). Subsequently, MOLLI-5(3)3 and MOLLI-4(1)3(1)2 were implemented to improve acquisition efficiency (3). Pixel-wise curve fitting algorithm using 3-parameter model is applied to estimate T1 values at each pixel. In this study, we sought to develop a fully-connected neural network (MyoMapNet)-based rapid myocardial T1 mapping approach using a single LL experiment with scan duration of 4-5 heartbeats and near-instantaneous (~2ms) reconstruction time.Methods
The data acquisition for MyoMapNet consists of collecting 5 (native) or 4 (post-contrast) T1 weighted images after application of an inversion pulse. Images are collected using a single-shot acquisition during the quiescent period in mid-diastole. Figure 1 shows the MyoMapNet architecture for T1 estimation from T1-weighted images. MyoMapNet is a fully connected neural network that consists of input, output, and 5 hidden layers. The input layer has 2Nt nodes for Nt T1 weighted signals and their corresponding inversion times, where Nt=5 or 4 in native or post-contrast T1 networks, respectively. The number of nodes in the five hidden layers are 400, 400, 200, 200, and 100, respectively, with a leaky-ReLU after each hidden layer. Stochastic gradient descent optimizer with learning-rate of 1E-8 and momentum of 0.8 was used to train the network for 2000 epochs with batch-size of 80 maps.Simulation experiments: Simulated MOLLI-5(3)3 dataset (80,000 samples) was generated and divided into training (80%; 64,000 samples) and testing (20%; 16,000 samples) signal-intensity time courses. Simulated T1 ranged from 400ms to 2000ms with an increment of 0.1 ms, T2 of 42 ms, and a fixed heart-rate of 60 bpm. Different Gaussian noise levels were added to the simulated signals for SNR of 20, 40, and 100.
In-vivo experiments: T1 mapping using MOLLI was collected in 717 subjects (386 males, 55±16.5 years old) with a subset of 535 subjects (232 male, 56.5±15 years old) with both native and post-contrast T1 maps using 3T scanner (MAGNETOM Vida, Siemens, Erlangen, Germany). All data were extracted retrospectively from clinical patients who were referred for a clinical CMR exam. Study protocol was approved by the Institutional Review Board and written consent was waived. MOLLI-5(3)3 and MOLLI-4(1)3(1)2 T1 was calculated off-line using 3-parameter curve-fitting model. Training datasets of 573 patients (1719 native T1 maps) and 428 patients (1281 post-contrast T1 maps) were randomly selected for native and post-contrast maps reconstruction, respectively. The performance of MyoMapNet was evaluated using a testing dataset of native T1 maps from 144 patients (432 MOLLI-5(3)3 T1 maps) and post-contrast T1 maps from 107 patients (324 MOLLI-4(1)3(1)2 T1 maps).
Data Analysis: For each T1 map, we calculated 3 sets of T1 estimates: (a) standard MOLLI T1 mapping using all collected images (8, or 9 images in native or post-contrast T1 mapping, respectively), (b) abbreviated MOLLI using only 5 native (MOLLI-5) or 4 post-contrast MOLLI (MOLLI-4) images using 3-parameter curve-fitting, and (c) MyoMapNet using only 5 native or 4 post-contrast MOLLI images. The ECV values were calculated for 75 patients who had both native and post-contrast T1 mapping in the testing dataset.
Results
Figure 2 shows Bland-Altman plots of MOLLI-5 and MyoMapNet at different SNR levels of simulated data. For higher SNR, MOLLI-5 exhibits systematic errors as a function of T1 values while MyoMapNet shows no systematic error. As SNR decreases, MyoMapNet significantly reduce the estimation errors compared to MOLLI-5 (0±15ms vs. -3±32ms; p-value<0.001) at SNR=40.Figure 3 shows example native T1 maps reconstructed with the MOLLI-5(3)3, MOLLI-5, and MyoMapNet in three subjects. MyoMapNet showed sharp myocardial T1 boundaries with more homogeneous T1 estimations than MOLLI-5 method. MyoMapNet was less sensitive to motion artifacts in cases with breath-holding failure (Figure 4).
In patient data, the mean T1 values estimated by MyoMapNet was similar to standard MOLLI methods (1200±45ms and 1199±46ms; p-value=0.3) in native T1, (563±45ms and 565±47ms; p-value=0.07) in post-contrast T1, and (27±4% and 27±4%; p-value=0.4) in ECV values. MyoMapNet estimated T1 values had smaller error than MOLL-5 (1±17ms vs. 31±34ms, p-value<0.001) in native T1, (-3±18ms vs. -17±23ms, p-value<0.001) for post-contrast, and (0.1±1.3% vs. 1.9±2.5%, p-value<0.001) for ECV values (Figure 5).
The average time for estimating T1 per map was ~25sec by a 3-parameter fitting model without motion correction using Matlab on CPU and 2ms by MyoMapNet using Python on GPU.
Conclusion
MyoMapNet enables fast and precise T1 mapping quantification from only 4-5 T1-weighted images, leading to shorter scan time and breath-holds of 4-5 heartbeats.Acknowledgements
No acknowledgement found.References
1. Messroghli DR, Moon JC, Ferreira VM, et al. Clinical recommendations for cardiovascular magnetic resonance mapping of T1, T2, T2* and extracellular volume: A consensus statement by the Society for Cardiovascular Magnetic Resonance (SCMR) endorsed by the European Association for Cardiovascular Imaging (EACVI). J. Cardiovasc. Magn. Reson. 2017;19:75.2. Messroghli DR, Radjenovic A, Kozerke S, Higgins DM, Sivananthan MU, Ridgway JP. Modified Look-Locker inversion recovery (MOLLI) for high-resolution T1 mapping of the heart. Magn. Reson. Med. 2004;52:141–146.
3. Kellman P, Hansen MS. T1-mapping in the heart: accuracy and precision. J. Cardiovasc. Magn. Reson. 2014;16:2.
Figures
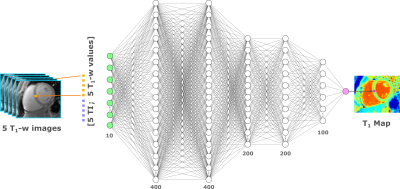
Figure 1. MyoMapNet architecture: MyoMapNet uses a fully-connected neural network for estimating voxel-wise
T1 values from T1-weighted images collected after a single
look-locker inversion pulse. For each voxel, the signal values from 5 T1-weighted images are concatenated
with their corresponding look-locker times and used as the network input (i.e.
10×1) for native T1 mapping. The input values are fed to a
fully-connected network with 5 hidden layers with 400, 400, 200, 200, and 100
nodes each layer, respectively. The output is the estimated T1 value
at each voxel.
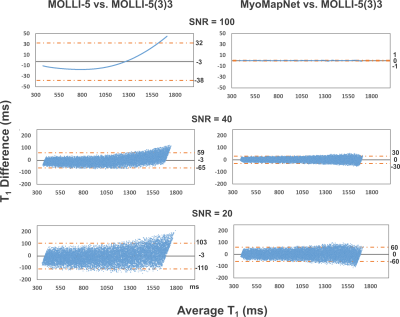
Figure 2. Bland-Altman plots showing the mean difference and 95% limits of
agreement between the simulated T1 values by the MOLLI-5 vs.
MOLLI-5(3)3 (left column) and MyoMapNet vs. MOLLI-5(3)3 (right column) for SNR
of 100, 40, and 20. MOLLI-5 yields a systematic error in T1
estimations, which is corrected in MyoMapNet.
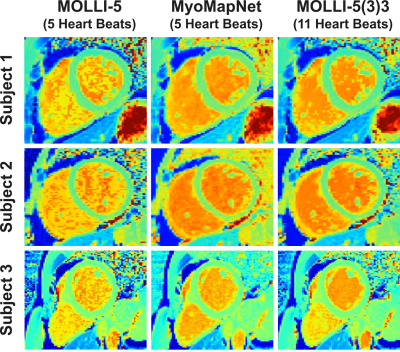
Figure 3. Native T1 maps from three patients, reconstructed using MOLLI-5
(using only 5 T1 weighted images with 3-parameter fitting),
MyoMapNet, and MOLLI-5(3)3 with a 3-parameter fitting model. MoyMapNet yield
maps with more homogenous signal compared to MOLLI-5.
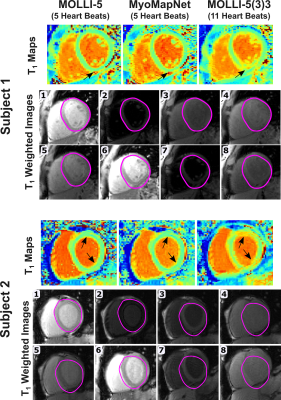
Figure 4. Myocardial T1 maps calculated using MOLLI-5 with a
3-parameter fitting model, MyoMapNet, and MOLLI-5(3)3, demonstrating the impact
of respiratory motion (black arrows) in two subjects. The epicardial
delineation of image #1 was copied to all other T1-weighted images
to show the degree of motion between images. Both subjects were instructed to
hold their breath during the scan.
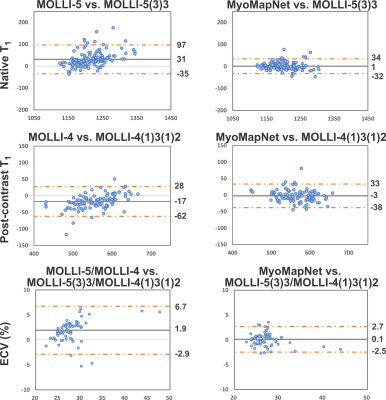
Figure 5. Bland-Altman plots showing the mean difference and 95% limits of
agreement for global native, post-contrast T1, and ECV for the
abbreviated MOLLI-5/MOLLI-4 vs. standard MOLLI-5(3)3/MOLLI-4(1)3(1)2 (left
column) and MyoMapNet vs. standard MOLLI-5(3)3/MOLLI-4(1)3(1)2 (right column).
Each dot represents the values calculated from one patient.