0442
A regularized joint water/fat separation and B0 map estimation for 2D-navigated interleaved EPI based diffusion MRI1C.J. Gorter Center for High Field MRI, Department of Radiology, LUMC, Leiden, Netherlands, 2Division of Image Processing, Department of Radiology, LUMC, Leiden, Netherlands, 3University of Lübeck, Lübeck, Germany, 4Philips Research Hamburg, Hamburg, Germany
Synopsis
Multi-shot EPI enables high-resolution diffusion-weighted imaging with reduced geometric distortions. However, fat is often a confounding factor in EPI, especially in regions with severe B0 inhomogeneities. For the proposed method, data is acquired using TE-shifted interleaved EPI and 2D-navigators to sense the motion-induced shot phases. The reconstruction includes a multi-peak fat model and corrects for the fat frequency-specific chemical shift displacements in phase-encoding direction by a time-efficient image-space formulation. In-vivo results show that the proposed algorithm provides improved water/fat separation compared with the conventional technique and fat-suppressed acquisition.
Introduction
Diffusion-weighted imaging (DWI) is an important contrast in diagnosing and staging of tumors1,2 which is often based on single-shot EPI (ssh-EPI). In comparison, multi-shot EPI (msh-EPI) with a 2D-navigator to sense the physiological motion has gained popularity due to the shorter echo train length and reduced geometric distortions3,4. Another common issue for EPI-based DWI is the presence of fat signal that has large spatial displacements in phase-encoding direction. Conventionally, spectral selective techniques such as SPIR5 have been used to suppress fat signals. However, those are prone to fail in regions of high B0 inhomogeneity6. Thus, water/fat separation via chemical-shift encoding might become an effective option7. Moreover, due to the fat spectrum’s multipeak profile, fat signals with different resonance frequencies will be spatially shifted to different locations in EPI images. Currently, researches attempted to address these challenges by a combination of chemical-shift encoding and spectral selective fat suppression approaches8,9. The k-space based algorithm could be another alternative but might suffer from an impractical long reconstruction time10. Tailored to the EPI, we propose a joint regularized algorithm with a series of shift matrices in image space to time-efficiently estimate accurate water, fat and B0 maps. In-vivo data were acquired through a TE-shifted navigator-based DW msh-EPI to enable chemical-shift encoding.Methods
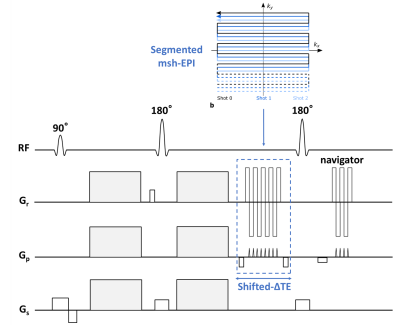
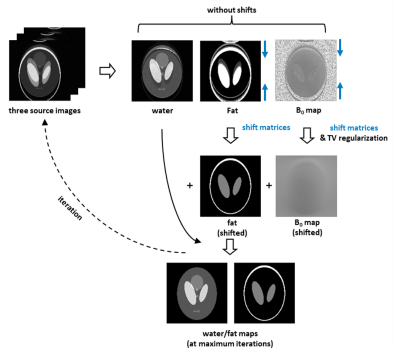
A custom 2D-navigated DW msh-SE-EPI sequence segmented along the phase-encoding direction was complemented to enable time-shifts of the msh-EPI sampling window, depicted in Fig. 1. After data acquisition, N = 3 chemical-shift encoded source images are obtained through IRIS4 reconstruction to correct motion-induced physiological phase changes. The different spatial shifts along the phase-encoding direction of the fat components compared to the water signal and the local B0 value are explicitly taken into account in the signal model:$$S_{n}\left(w(x,y),f(x,y),\varphi_{B}(x,y)\right)=w(x,y)e^{i2\pi\varphi_{B}(x,y)\Delta\mathrm{TE}_{n}}+\sum_{m=1}^{M}\alpha_{m}f\left(x,y-\Delta y_{m}\right)e^{i2\pi\varphi_{B}\left(x,y-\Delta y_{m}\right) \Delta\mathrm{TE}_{n}}e^{i2 \pi\varphi_{F, m}\Delta\mathrm{TE}_{n}}+v_{n}(x, y)$$where $$$w$$$ and $$$f$$$ are the magnitudes of water and fat signals, $$$\Delta\mathrm{TE}_{n}$$$[s] the $$$n$$$-th TE-shifted scan, $$$\varphi_{B}$$$[Hz] the B0 field at certain $$$(x,y)$$$ and originated $$$(x,y-\Delta y_{m})$$$ locations in phase-encoding direction. Moreover, $$$\alpha_{m}$$$ and $$$\varphi_{F, m}$$$ [Hz] denote the relative weight and resonance frequency for each fat peak $$$m$$$, and $$$v_{n}(x, y)$$$ is complex noise. For water/fat separation, the signal equation poses the following minimization problem:$$\{W,F\}=\underset{\widehat{W},\widehat{F}\in\mathbb{C}^{Q}}{\operatorname{argmin}}\|\hat{A}X-S\|_{2}^{2}$$Where $$$Q$$$ is the number of voxels.$$$S=\left[s_{1}^{1},\ldots,s_{1}^{Q},\ldots,s_{N}^{1},\ldots,s_{N}^{Q}\right]^{T}$$$ and $$$X=[W,F]^{T}=\left[w^{1},\ldots,w^{Q},f^{1},\ldots,f^{Q}\right]^{T}$$$ are vectorized representations of the $$$N$$$ source images and water/fat contents. $$$\hat{A}$$$ is the joint coefficient matrix with two series of shift matrices acting on fat signals and fat associated B0 map to shift them back to the original location. The B0 map can be estimated by Gauss-Newton optimization:$$\left\{\Delta\Phi_{B},\Delta W,\Delta F\right\}=\underset{\Delta\varphi_{B}\in\mathbb{R}^{Q}\\ \Delta w,\Delta f\in\mathbb{C}^{Q}}{\operatorname{argmin}}\|\Delta S-\hat{B}\Delta Y\|_{2}^{2}+\lambda TV\left(\Delta\Phi_{B}\right)$$where $$$\Delta Y=\left[\Delta W,\Delta F,\Delta\Phi_{B}\right]^{T}=\left[\Delta w^{1},\ldots,\Delta w^{Q},\Delta f^{1},\ldots,\Delta f^{Q},\Delta \varphi_{B}^{1},\ldots,\Delta\varphi_{B}^{Q}\right]^{T}$$$ and $$$\hat{B}$$$ is the coefficient matrix of the Gauss-Newton search system with shift matrices to shift back $$$\Delta F$$$ and $$$\Delta\Phi_{B}$$$ for each individual peak. $$$TV$$$ is the total variation regularization operator to enforce smoothness of the updated field map error $$$\Delta\Phi_{B}$$$ for the next iteration. Calculation of the two least-square systems are repeated (Fig.2) until the maximum of iterations (e.g.,6) is reached. In addition, for anatomies with severe field inhomogeneities (e.g., head/neck regions), an additional extrapolation step as used for coil-sensitivity maps11 is adopted after half-maximum iterations to give a better initialization for the remaining iterations. A self-calibrated 7-peak fat model based on Ren et al.12, but at the appropriate TE, was applied13. A conventional multipeak IDEAL6,14 algorithm using the same fat spectrum model was implemented for comparison.Experiments were conducted in the leg and the head/neck region with 5 subjects on a 3T scanner (Philips, Best, The Netherlands). Each spin-echo DWI experiment was performed with three b-values (0, 300, 600 s/mm2), 6/8 interleaves, TE = 59/53 ms, TR = 2000 ms and at two resolutions: 1.44 x 1.53 x 4 mm3/1.19 x 1.23 x 4 mm3 with acquisition matrices 152 × 160/168 x 162. The three source images were acquired at 0.2/1.0/1.8 ms with respect to the spin echo. For the given matrix sizes, the estimation required 14.9/15.6 s per iteration. In addition, for each measurement another DWI scan was acquired using conventional SPIR for comparison.
Results and Discussion
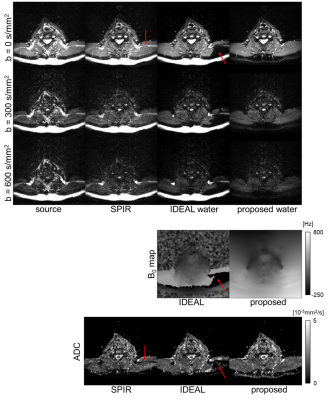
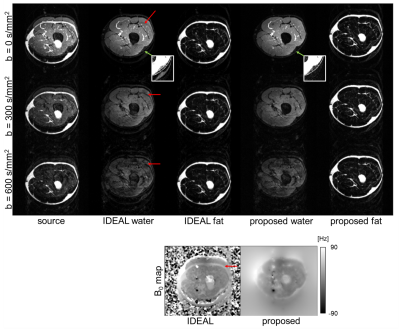
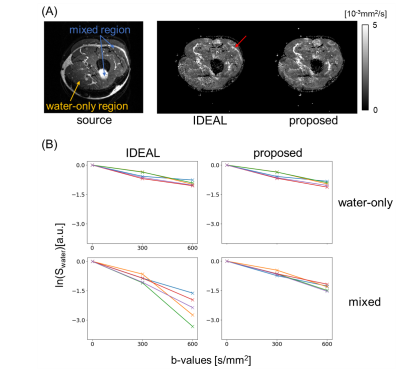
Figure 3 illustrates the DWI results of one subject’s head-neck images of SPIR image and water/B0 maps estimated from IDEAL/proposed algorithms with ADC calculations. For both SPIR and IDEAL, fat suppression failed in areas with large field inhomogeneities while this was not the case for our proposed algorithm. Figure 4 and 5 show results from IDEAL and our proposed algorithm for one subject’s leg. In Fig.4, the rim-like artifact observed with IDEAL can be eliminated by the proposed algorithm. In Fig.5, the water-only and mixed regions (both water/fat present) were segmented in one source image. The result of the proposed algorithm shows good linearity of the natural logarithm of DWI signals among 5 pixels in both regions, while IDEAL shows nonlinear behaviour in mixed region.Conclusion
The proposed algorithm provides more reliable water/fat separation and diffusion quantification compared to standard techniques. Future work will include undersampling to support accelerated data acquisition. To further enhance the image quality, fat signal present in 2D-navigators is considered to be eliminated through further approaches15.Acknowledgements
The authors would like to acknowledge NWO-TTW (HTSM-17104) for funding this project.References
1. Le Bihan D, Mangin J-F, Poupon C, et al. Diffusion tensor imaging: concepts and applications. JMRI. 2001;13(4):534–546.
2. Wu W, Miller KL. Image formation in diffusion MRI: A review of recent technical developments: Review of Image Formation in dMRI. JMRI. 2017;46(3):646-662.
3. McKinnon GC. Ultrafast interleaved gradient-echo-planar imaging on a standard scanner. Magn Reson Med. 1993;30(5):609-616. doi:10.1002/mrm.1910300512
4. Jeong H-K, Gore JC, Anderson AW. High-resolution human diffusion tensor imaging using 2-D navigated multishot SENSE EPI at 7 T. MRM. 2013;69(3):793-802. doi:10.1002/mrm.24320
5. Zee CS, Segall HD, Terk MR, et al. SPIR MRI in spinal diseases. J Comput Assist Tomogr 1992;16:356-360.
6. Anzai Y, Lufkin RB, Jabour BA, Hanafee WN. Fat-suppression failure artifacts simulating pathology on frequency-selective fat-suppression MR images of the head and neck. AJNR Am J Neuroradiol. 1992 May-Jun;13(3):879-84. PMID: 1590186.
7. Reeder, S. B., Wen, Z., Yu, H., Pineda, A. R., Gold, G. E., Markl, M., & Pelc, N. J. (2004). Multicoil Dixon Chemical Species Separation with an Iterative Least-Squares Estimation Method. Magnetic Resonance in Medicine, 51(1), 35–45. https://doi.org/10.1002/mrm.10675
8. Hernando D, Karampinos DC, King KF, Haldar JP, Majumdar S, Georgiadis JG, Liang Z‐PP. Removal of olefinic fat chemical-shift artifact in diffusion MRI. Magn Reson Med 2011;65:692–701.
9. Burakiewicz, J., Hooijmans, M.T., Webb, A.G., Verschuuren, J.J., Niks, E.H. and Kan, H.E. (2018), Improved olefinic fat suppression in skeletal muscle DTI using a magnitude‐based dixon method. Magn. Reson. Med., 79: 152-159. doi:10.1002/mrm.26655
10. Honorato, J.L., Parot, V., Tejos, C., Uribe, S. and Irarrazaval, P. (2012), Chemical species separation with simultaneous estimation of field map and T2* using a k-space formulation. Magnetic Resonance Medicine, 68: 400-408. doi:10.1002/mrm.23237
11. Liu, C., Bammer, R., & Moseley, M. E. (2007). Parallel imaging reconstruction for arbitrary trajectories using k-space sparse matrices (kSPA). Magnetic Resonance in Medicine, 58(6), 1171–1181. https://doi.org/10.1002/mrm.21334
12. Ren J, Dimitrov I, Sherry AD, Malloy CR. Composition of adipose tis- sue and marrow fat in humans by 1H NMR at 7 Tesla. J Lipid Res 2008;49:2055–2062.
13. Yu H, Shimakawa A, McKenzie CA, Brodsky E, Brittain JH, Reeder SB. Multiecho water-fat separation and simultaneous R2* estimation with multifrequency fat spectrum modeling. Magn Reson Med. 2008;60(5):1122-1134. doi:10.1002/mrm.21737
14. Reeder SB, Pineda AR, Wen Z, Shimakawa A, Yu H, Brittain JH, Gold GE, Beaulieu CH, Pelc NJ. Iterative decomposition of water and fat with echo asymmetry and least-squares estimation (IDEAL): application with fast spin-echo imaging. Magn Reson Med. 2005 Sep;54(3):636-44. doi: 10.1002/mrm.20624.
15. Gomori JM, Holland GA, Grossman RI, Gefter WB, Lenkinski RE. Fat suppression by section-select gradient reversal on spin-echo MR imaging. Work in progress. Radiology. 1988 Aug;168(2):493-5. doi: 10.1148/radiology.168.2.3393670. PMID: 3393670
Figures