0425
Mask R-CNN for Segmentation of Kidneys in Magnetic Resonance Imaging1Radiology, UT Southwestern Medical Center, Dallas, TX, United States
Synopsis
Automated segmentation of kidneys in Magnetic Resonance Imaging (MRI) exams are important for enabling radiomics and machine learning analysis of renal disease. In this work, we propose to use a deep learning method called Mask R-CNN for the segmentation of kidneys in 2D coronal T2W FSE images of 94 MRI exams. With 5-fold cross-validation data, the Mask R-CNN is trained and validated on 66 and 9 MRI exams and then evaluated on the remaining 19 exams. Our proposed method achieved an average dice score of 0.839 and an average IoU of 0.763.
Summary
This paper validated the Mask R-CNN for the segmentation of Kidneys in 2D T2-weighted fast spin-echo slices of 94 MRI exams. We achieved an average dice score of 83.9% and IoU of 76.3% in 5-fold cross-validation data.Introduction
Kidney cancer is among the 6th most common cancers in men and 8th in women, and the 5-year survival rate is 75% 1,2. However, there is wide variability in the prognosis among different renal cancers. Most kidney cancers (70%) are first diagnosed as an incidental small renal mass (SRM; ≤4 cm) on an imaging study performed for an unrelated clinical reason. Furthermore, many of these renal masses are benign tumors that mimic cancers. Although radiomics may improve the characterization of renal masses3, these analyses are often time-consuming. An automated deep learning system that provides automatic segmentation of kidney and renal masses would be a useful supporting tool for clinicians 3,4. In recent times, artificial intelligence (AI) and deep learning methods such as convolution neural networks (CNN) have achieved good results in automated recognition of abnormalities in various medical imaging modalities such as X-ray, computed tomography (CT), positron emission tomography (PET), and MRI 4-8. An AI segmentation tool for SRM is lacking. And reports about automatic segmentation of the kidneys on MRI with AI algorithms have focused primarily on patients with adult polycystic kidney disease 9. Moreover, respiratory motion between different MRI acquisitions and between slices of the same acquisition makes AI more challenging. We hypothesize that accurate segmentation of the normal renal parenchyma is the first step toward the development of fully automated AI algorithms for renal mass characterization. This work focuses on the validation of a Mask R-CNN for the automated segmentation of kidneys in 2D T2-weighted (T2w) fast spin-echo (FSE) images of MRI exams.Dataset
Our dataset consists of 94 MRI exams from patients with known renal masses imaged at different institutions and referred to our center for definitive treatment. We used the coronal 2D T2w FSE images of each MRI exam. The number of T2W slices per patient varied from 11 to 56, and the total number of 2D images was 2423. We first pre-processed the MRI exams with the N4 bias correction algorithm. All images were annotated by a third-year radiology resident who created kidney masks per patient. These masks were used as ground truth in this study.Mask R-CNN Architecture
Mask R-CNN is a deep learning architecture extended from Faster R-CNN, which determines the object's location by drawing a bounding box (detection) and then marking each pixel of the object with a mask (segmentation) 12. In this work, we used the Mask R-CNN architecture with InceptionResNetV2 as the CNN network to segment kidneys in 2D T2W FSE images. We used the TensorFlow object detection library to implement the Mask R-CNN InceptionResNetV2 method with 32 GB Nvidia Titan V100 GPU. We set a batch size of 4, the total number of epochs is 100, an initial learning rate of 0.008, with a momentum optimizer of 0.9, and gradient clipping by norm of 10. The final evaluation model is selected based on minimum validation loss. We also used the pre-trained model for fine-tuning and data augmentation techniques such as horizontal and vertical flips on the fly during training.Results and Discussion
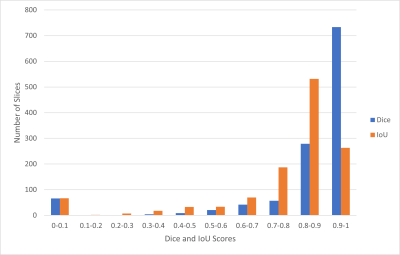
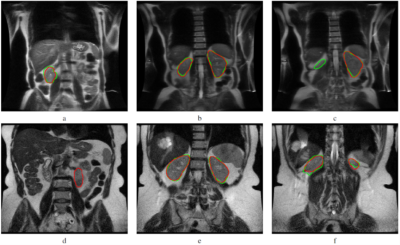
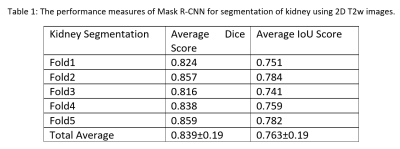
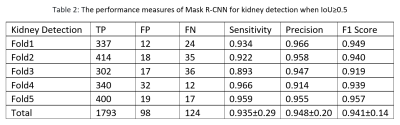
The Mask R-CNN method was trained on approximately 66 MRI exams as a training set and validated on nine exams, and the remaining 19 exams were used as a testing set in a single fold. We evaluated the performance of Mask R-CNN on 5-fold cross-validation data to test the whole dataset. Our proposed method achieved the average dice score of 0.839 with a standard deviation (±) of 0.19 and IoU of 0.763±0.19 across all folds, as shown in Table 1. The distribution of the number of slices in which dice and IoU scores ranged from 0 to 1 is shown in Fig. 1. The low standard deviation (SD) suggests, the method performs consistently well across all the folds. We further evaluated the kidney detection by setting the IoU≥0.5 to determine the true positive (TP), false positive (FP), false negative (FN), sensitivity, precision and F1 score. The performance metrics of kidney detection are shown in Table 2. Our method achieved an average sensitivity of 0.935±0.29, precision of 0.948±0.20, and F1 score of 0.941±0.14. The method detected the shape and location of the kidney correctly. A few examples of the correct and incorrect inferences are shown in Fig. 2.Conclusion
In this work, we validated the Mask R-CNN for the segmentation of kidneys in 2D coronal T2W FSE images. Our method performed consistently well in all performance measures for 5-fold cross-validation data. Our proposed method accurately segmented the kidney, hence, provides us an automated application to assess the location and size of the kidney in MRI exams. It is also an essential step in applying textural analysis radiomics where the kidney segmentation is required to aid in the diagnosis of abnormalities 11, 12. The main limitation of this work is the small dataset. Further studies will be conducted to validate in larger datasets and other MRI sequences such as T1-weighted.Acknowledgements
No acknowledgement found.References
1. “Key Statistics About Kidney Cancer,” American Cancer Society. [Online]. Available: https://www.cancer.org/cancer/kidney-cancer/about/key-statistics.html. [Accessed: 14-Dec-2020]
2. Kidney cancer. (2019, July 9). Centers for Disease Control and Prevention. https://www.cdc.gov/cancer/kidney/index.html [Accessed: 14-Dec-2020]
3. Dwivedi, D. K., Xi, Y., Kapur, P., Madhuranthakam, A. J., Lewis, M. A., Udayakumar, D., ... & Fulkerson, M. (2020). Magnetic Resonance Imaging Radiomics Analyses for Prediction of High-Grade Histology and Necrosis in Clear Cell Renal Cell Carcinoma: Preliminary Experience. Clinical Genitourinary Cancer.
4. Souza, J. C., Diniz, J. O. B., Ferreira, J. L., da Silva, G. L. F., Silva, A. C., & de Paiva, A. C. (2019). An automatic method for lung segmentation and reconstruction in chest X-ray using deep neural networks. Computer methods and programs in biomedicine, 177, 285-296.
5. Tomita, N., Cheung, Y. Y., & Hassanpour, S. (2018). Deep neural networks for automatic detection of osteoporotic vertebral fractures on CT scans. Computers in biology and medicine, 98, 8-15.
6. Goyal, M., Oakley, A., Bansal, P., Dancey, D., & Yap, M. H. (2019). Skin lesion segmentation in dermoscopic images with ensemble deep learning methods. IEEE Access, 8, 4171-4181.
7. Hatt, M., Laurent, B., Ouahabi, A., Fayad, H., Tan, S., Li, L., ... & Drapejkowski, F. (2018). The first MICCAI challenge on PET tumor segmentation. Medical image analysis, 44, 177-195.
8. Akkus, Z., Galimzianova, A., Hoogi, A., Rubin, D. L., & Erickson, B. J. (2017). Deep learning for brain MRI segmentation: state of the art and future directions. Journal of digital imaging, 30(4), 449-459.
9. Kline, T. L., Korfiatis, P., Edwards, M. E., Blais, J. D., Czerwiec, F. S., Harris, P. C., ... & Erickson, B. J. (2017). Performance of an artificial multi-observer deep neural network for fully automated segmentation of polycystic kidneys. Journal of digital imaging, 30(4), 442-448.
10. He, K., Gkioxari, G., Dollár, P., & Girshick, R. (2017). Mask r-cnn. In Proceedings of the IEEE international conference on computer vision (pp. 2961-2969).
11. de Leon, A. D., Kapur, P., & Pedrosa, I. (2019). Radiomics in kidney Cancer: MR imaging. Magnetic Resonance Imaging Clinics, 27(1), 1-13. 12. Krajewski, K. M., & Pedrosa, I. (2018). Imaging advances in the management of kidney cancer. Journal of Clinical Oncology, 36(36), 3582.
Figures