0393
Image domain Deep-SLR for Joint Reconstruction-Segmentation of Parallel MRI1Electrical and Computer Engineering, The University of Iowa, Iowa City, IA, United States
Synopsis
We present a novel framework for the joint reconstruction and segmentation of parallel MRI (PMRI) brain data. We introduce an image domain deep network for calibrationless recovery of undersampled PMRI data. It is deep-learning based generalization of local low-rank approaches for uncalibrated PMRI recovery including CLEAR. The image domain approach exploits additional annihilation relations compared to k-space based approaches and hence offers improved performance. To minimize segmentation errors resulting from undersampling artifacts, we combined the proposed scheme with a segmentation network and trained it end-to-end. It offers improved reconstruction with reduced blurring and sharper edges than independently trained reconstruction network.
Introduction
Modern MRI methods often rely on acceleration methods including Parallel MRI (PMRI) and compressive sensing to reduce the scan time. Calibrated methods such as SENSE/GRAPPA [1, 2] as well as calibrationless structured low-rank methods [3, 4] have been introduced to recover the images from undersampled measurements. Our recent work has shown that linear relations between their k-space measurements can be capitalized using k-space deep-learning (DL) strategies [5],which are more computationally efficient than classical methods. Despite the great progress made in image reconstruction, undersampling artifacts and blurring of image edges are inevitable at high acceleration factors; these artifacts can deteriorate the performance of segmentation algorithms that exploit the edges and the coherence of image intensities within regions that would be impacted by undersampling.We introduce a novel framework for DL based calibration-free MRI reconstruction and segmentation. The contributions include a novel image domain deep structured low-rank framework for calibration-free PMRI recovery and a joint segmentation-reconstruction framework to minimize segmentation errors introduced by undersampling artifacts and improve reconstruction quality.
Proposed Method
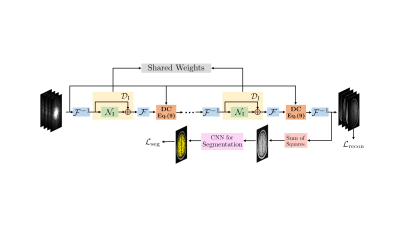
The forward model is defined as, \begin{equation} \mathbf b_i = \underbrace{\mathcal S \mathcal F}_{\mathcal A} ~\gamma_i + \mathbf n_i, \hspace{2pt} i = 1 \ldots N \end{equation} where $$$\mathbf b_i$$$ are noisy Fourier coefficients of $$$i^{th}$$$ coil image $$$\gamma_i$$$ corrupted by Gaussian noise $$$\mathbf n_i$$$, $$$N$$$ is the total number of coils, $$$\mathcal F$$$ is the Fourier operator and $$$\mathcal S$$$ denotes the sampling operator. According to the CLEAR formulation [6], if $$$P_{\mathbf s_0}$$$ is a patch extraction operator that extracts $$$M\times M$$$ patches of $$$\gamma_i(\mathbf s)$$$ centered at $$$\mathbf s_0$$$, the matrices \begin{equation}\label{key} \boldsymbol\Gamma_{\mathbf s} = \begin{bmatrix} P_{\mathbf s}(\gamma_1)| & \ldots & |P_{\mathbf s}(\gamma_N) \end{bmatrix} \end{equation} are low-rank. It solves for $$$\Gamma = \begin{bmatrix} \gamma_1,.. ,\gamma_N \end{bmatrix}$$$ as the nuclear norm minimization problem \begin{equation}\label{clear} \boldsymbol \Gamma = \arg \min_{\boldsymbol \Gamma} \|\mathcal A(\boldsymbol\Gamma)-\mathbf B\|^2 + \lambda\sum_s \|\boldsymbol\Gamma_{\mathbf s}\|_* \end{equation}.Using the iterative reweighted least squares formulation, we obtain a model-based DL network alternating between \begin{eqnarray} \mathbf X_{n} &=& \mathcal D_{\rm I}(\boldsymbol \Gamma_n)\\ \widehat{\boldsymbol \Gamma}_{n+1} &=& (\mathcal A^H \mathcal A + \lambda \mathbf I)^{-1}(\mathcal A^H \mathbf B + \lambda \widehat{\mathbf X}_{n}) \end{eqnarray}
Similar to [5], the network is unrolled and trained end-to-end with exemplar data with CNN weights shared across iterations. The training loss is $$$\mathcal L_{\rm recon} = \|\boldsymbol \Gamma - \boldsymbol \Gamma_{gs} \|^2$$$ , where $$$\boldsymbol\Gamma$$$ is the prediction and $$$\boldsymbol \Gamma_{gs}$$$ is the gold standard multicoil data obtained from fully sampled measurements. The proposed method I-DSLR eliminates the need for calibration data for estimating the coil sensitivities or linear filters $$$\mathbf Q_s$$$.
We propose a multi-task deep network as shown in Fig .1, trained end-to-end. A segmentation network is attached to the final iteration of the I-DSLR. We use a weighted linear combination of normalized mean squared error $$$\mathcal L_{\rm recon}$$$ and pixel-wise multi-label cross entropy $$$\mathcal L_{\rm seg} = -\sum_p \boldsymbol{\phi}_{p}^{gs} \ln \boldsymbol{\phi}_{p}$$$ error for training. \begin{equation} \mathcal L_{\rm total} = \mathcal L_{\rm recon} + \beta \mathcal L_{\rm seg} \end{equation} For each pixel $$$p$$$, $$$\boldsymbol{\phi}_{gs}$$$ is the gold standard segmentation on the sum-of-squares image obtained from $$$\boldsymbol{\Gamma}_{gs}$$$ and $$$\boldsymbol{\phi}$$$ is the segmentation CNN output.
Experiments and Results
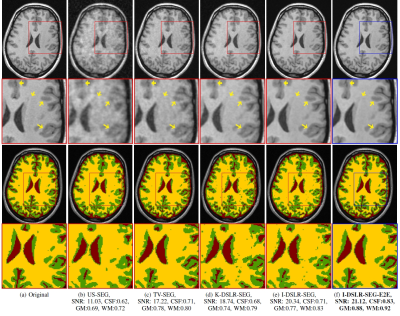
We perform experiments on the publicly available Calgary Campinas Dataset (CCP) [7]. It consists of 12-channel raw k-space data of T1-weighted brain MRI scans from a Discovery MR750 3T scanner for 67 subjects. The slice dimensions are 208 x 170 for axial view of the brain. Ground truth segmentations for the slices were generated using the FAST software. Forty subjects (40 x 256 = 10240 slices) were used for training, 7 for validation and the remaining 20 for testing purposes. 2D non-uniform cartesian variable density undersampling masks with different acceleration factors were used for experiments; readout direction is orthogonal to axial slices.A pre-trained I-DSLR network cascaded with a segmentation network pre-trained using fully sampled data (I-DSLR-SEG) is compared against k-space Deep-SLR method(K-DSLR-SEG), total variation (TV-SEG) and undersampled (US-SEG) in the same setting. We also compare against I-DSLR-SEG-E2E, a cascade of IDSLR and UNET segmentation networks trained end-to-end (E2E). In TV-SEG, the segmentation network is trained and tested on images reconstructed using TV. Similarly, for US-SEG, the segmentation network is trained and tested on undersampled datasets.
A comparison of the methods is shown in Fig. 2. I-DSLR has sharper edges compared to US, TV and K-DSLR. The corresponding segmentation performance improves with increase in reconstruction quality. The end-to-end training strategy in I-DSLRSEG-E2E further improves quality over I-DSLR-SEG. Its improved reconstruction can be attributed to the regularization by the segmentation network and vice-versa.
Conclusion
We introduced a novel image domain model-based DL approach for calibrationless PMRI recovery. It is a non-linear extension of locally low rank methods for calibrationless parallel MRI. The experiments show that additional annihilation relations exploited by I-DSLR approach offers better performance over k-space approach K-DSLR [5]. We introduce a multi-task framework where I-DSLR is cascaded with a segmentation dedicated DL network which is trained end-to-end. The networks regularize each other, thereby reducing the errors caused from undersampling artifacts. Experiments show that the segmentation accuracy depends on the reconstruction quality. I-DSLR-SEG-E2E reduces the errors propagating to segmentation network, thus outperforming methods that cascade independently trained networks.Acknowledgements
This work is supported by NIH 1R01EB019961-01A1.References
1. Pruessmann et al., “SENSE: sensitivity encoding for fast mri,” MRM, vol. 42, no. 5, pp. 952–962,1999.
2. Griswold et al., “Generalized autocalibrating partially parallel acquisitions (GRAPPA),” MRM, vol. 47, no. 6, pp. 1202–1210, 2002.
3. Uecker et al., “ESPIRiT—an eigenvalue approach to autocalibrating parallel MRI: where SENSE meets GRAPPA,” MRM, vol. 71, no. 3, pp. 990–1001, 2014.
4. Mathews et al., “Structured Low-Rank Algorithms: Theory, Magnetic Resonance Applications, and Links to Machine Learning,” IEEE SPM, vol. 37, no. 1, pp. 54–68, 2020.
5. Pramanik et al., “Deep Generalization of Structured LowRank Algorithms (Deep-SLR),” IEEE TMI, vol. 39, no. 12, pp. 4186-4197, 2020.
6. Joshua D Trzasko and Armando Manduca, “CLEAR: Calibration-free parallel imaging using locally low-rank encouraging reconstruction,” in ISMRM, 2012, vol. 517.
7. Souza et al., “An open, multi-vendor, multi-field-strength brain mr dataset and analysis of publicly available skull stripping methods agreement,” NeuroImage, vol. 170, pp. 482–494, 2018.
Figures