0336
Track-To-Learn: A general framework for tractography with deep reinforcement learning
Antoine Théberge1, Christian Desrosiers2, Maxime Descoteaux1, and Pierre-Marc Jodoin1
1Faculté des Sciences, Université de Sherbrooke, Sherbrooke, QC, Canada, 2Département de génie logiciel et des TI, École de technologie supérieure, Montréal, QC, Canada
1Faculté des Sciences, Université de Sherbrooke, Sherbrooke, QC, Canada, 2Département de génie logiciel et des TI, École de technologie supérieure, Montréal, QC, Canada
Synopsis
Supervised machine learning algorithms have been proposed to learn tractography algorithms implicitly from data, without relying on hard-to-develop anatomical priors. However, supervised learning methods rely on labelled data that is very hard to obtain. To remove the need for such data but still leverage the expressiveness of neural networks, we introduce and implement Track-To-Learn, a general framework to pose tractography as a deep reinforcement learning problem. We show that competitive results can be obtained on known data and that the learned algorithms are able to generalize far better to new, unseen data, than prior supervised learning-based tractography algorithms.
Introduction
Recently, supervised machine learning (SL) solutions have been proposed to learn tractography algorithms directly from reference tractograms1. These solutions are able to exhibit high performance on training data without the need for strong anatomical priors and are able to exploit the structured shape of streamlines to disentangle fiber crossings, kissings, and more.Obtaining enough labelled data to train SL methods is no easy feat. Due to the in-vivo nature of tractography, datasets either come from phantoms, which can only ever be so similar to real human anatomy, or from segmentation, which is prone to high variability2,3. As such, very few datasets with ground-truth streamlines are publicly available today1,4. While SL offers an appealing alternative to classical tractography algorithms, their generalization capabilities have yet to be demonstrated and we argue that the very nature of dMRI and tractography calls for methods that do not rely on ground-truth data.
To leverage the power of machine learning without relying on hard-to-obtain ground-truth data, we propose Track-to-Learn: a framework that formulates the tractography problem as a reinforcement learning (RL)5 problem.
Methods
We define the environment as the diffusion volume the agent will perform tractography in. Fiber orientation distribution function (fODF) spherical harmonics (SH) coefficients at the streamline’s “head”, along with the WM mask value and the last four tracking steps act as the state sent to the agent, which outputs a new tracking step. The environment propagates the streamline, computes a new state and sends it back to the agent along with the associated reward. To promote a smooth streamline propagation aligned with the diffusion signal, we pose the reward function as the cosine-similarity between the tracking step and the underlying peaks extracted from the fODFs, multiplied by the alignment between the new tracking step and the previous (c.f Figure 1 for the different components).We trained agents with two deep RL algorithms: TD36 and SAC7. In our implementation, both use three-layer feed-forward neural networks with a width of 1024 or 2048, depending on the experiment.
In experiment 1, we trained and evaluated the performance of our agents on a synthetic recreation of the FiberCup dataset8,9, which has 3 slices acquired at a 3mm iso resolution with b=1000 for 30 directions. To test their generalization capabilities, we flipped the FiberCup horizontally and tested on it without re-training. We compare our reconstruction and generalization capabilities against classical tractography algorithms, as well as Learn-to-Track10, an SL algorithm for tractography.
Second, we compared the performance of our method against prior work on the ISMRM2015 WM Tractography dataset11, by training and testing on the same dataset, as did prior methods. The ISMRM2015 dataset consists of 25 ground-truth bundles which were used to generate a synthetic b=1000 32 directions 2mm isotropic diffusion volume and matching T1 image. We report results for our agents, several prior methods and classical tractography algorithms.
Finally, we assessed the generalization capabilities of our agents by training them on a single HCP12 subject (ID 100206, 1.25mm isotropic diffusion volume, b values of 1000, 2000, 3000 with 90 directions each and 18 b0 images) and then testing on ISMRM2015. Although the HCP dataset offers no ground-truth, we could still perform training on it due to the unsupervised nature of our method. We report scores for our agents as well as prior methods.
Datasets for Experiment 2 and 3 were pre-processed using Tractoflow13, and all scores were reported by the Tractometer14. In experiment 1 and 3, Learn-to-Track10 was re-trained by the authors.
Results
Figure 2 provides a visual comparison of the reconstructions by the proposed method and by a prior SL method for both FiberCups. Figure 3 presents the results of the first experiment. Our method achieves competitive performance compared to previous methods on the original FiberCup. However, we can observe that the performance of SL methods degrades significantly on the “flipped” dataset, while ours does not.Figure 4 presents the results of our 2nd experiment showing that our method is highly competitive. While previous SL methods exhibit high performance when trained on the ground-truth data (which is never available on real in-vivo data), we see a clear drop in their performance when trained on manually segmented tractograms.
Figure 5 presents the results of our third experiment. Overall, our SAC agent outperforms or is competitive compared to all other methods .
Discussion & Conclusion
Results from experiment 1 and 2 show that the proposed framework is able to achieve highly competitive results without the need for labelled data. Moreso, prior work (both classical and machine learning-based) typically had to implement explicit priors to reduce their NC rate while the nature of the RL objective means that trained agents implicitly avoid early streamline termination. However, the proposed framework excels in its generalization capabilities: results from experiment 1 and 3 demonstrate that our method is able to achieve high performance when tracking on a different dataset that was used for training, as opposed to prior work.We argue that RL is best suited to tackle the tractography problem, and, through Track-to-Learn, have opened a brand new avenue of research that will hopefully lead to more representative tractography algorithms.
Acknowledgements
The authors would like to thank members of the SCIL and VITAL groups of the University of Sherbrooke for their suggestions, insight and discussions on this project.References
- Poulin, P., Jörgens, D., Jodoin, P. M., & Descoteaux, M. (2019). Tractography and machine learning: Current state and open challenges. Magnetic resonance imaging, 64, 37-48.
- Rheault, F., De Benedictis, A., Daducci, A., Maffei, C., Tax, C. M., Romascano, D., ... & Girard, G. (2020). Tractostorm: The what, why, and how of tractography dissection reproducibility. Human Brain Mapping, 41(7), 1859-1874.
- Schilling, Kurt G. et al. "Tractography dissection variability: what happens when 42 groups dissect 14 white matter bundles on the same dataset?". bioRxiv. (2020).
- Kurt G. Schilling, Alessandro Daducci, Klaus Maier-Hein, Cyril Poupon, Jean-Christophe Houde, Vishwesh Nath, Adam W. Anderson, Bennett A. Landman, & Maxime Descoteaux (2019). Challenges in diffusion MRI tractography – Lessons learned from international benchmark competitions. Magnetic Resonance Imaging, 57, 194 - 209.
- Sutton, R., & Barto, A. (2018). Reinforcement Learning: An Introduction. A Bradford Book.
- Fujimoto, S., Van Hoof, H., & Meger, D. (2018). Addressing function approximation error in actor-critic methods. arXiv preprint arXiv:1802.09477.
- Haarnoja, T., Zhou, A., Abbeel, P., & Levine, S. (2018). Soft actor-critic: Off-policy maximum entropy deep reinforcement learning with a stochastic actor. arXiv preprint arXiv:1801.01290.
- Poupon, C., Rieul, B., Kezele, I., Perrin, M., Poupon, F., & Mangin, J.F. (2008). New diffusion phantoms dedicated to the study and validation of high-angular-resolution diffusion imaging (HARDI) models. Magnetic Resonance in Medicine, 60(6), 1276-1283.
- Neher, P., Laun, F., Stieltjes, B., & Maier-Hein, K. (2014). Fiberfox: Facilitating the creation of realistic white matter software phantoms: Realistic White Matter Software Phantoms. Magnetic Resonance in Medicine, 72(5), 1460–1470.
- Poulin, P., Côté, M.A., Houde, J.C., Petit, L., Neher, P., Maier-Hein, K., Larochelle, H., & Descoteaux, M. (2017). Learn to Track: Deep Learning for Tractography. bioRxiv.
- Maier-Hein, K. H., Neher, P. F., Houde, J. C., Côté, M. A., Garyfallidis, E., Zhong, J., ... & Reddick, W. E. (2017). The challenge of mapping the human connectome based on diffusion tractography. Nature communications, 8(1), 1-13.
- Van Essen, D. C., Smith, S. M., Barch, D. M., Behrens, T. E., Yacoub, E., Ugurbil, K., & Wu-Minn HCP Consortium. (2013). The WU-Minn human connectome project: an overview. Neuroimage, 80, 62-79.
- Guillaume Theaud, Jean-Christophe Houde, Arnaud Boré, François Rheault, Felix Morency, & Maxime Descoteaux (2020). TractoFlow: A robust, efficient and reproducible diffusion MRI pipeline leveraging Nextflow & Singularity. NeuroImage, 218, 116889.
- Marc-Alexandre Côté, Gabriel Girard, Arnaud Boré, Eleftherios Garyfallidis, Jean-Christophe Houde, & Maxime Descoteaux (2013). Tractometer: Towards validation of tractography pipelines. Medical Image Analysis, 17(7), 844 - 857.
- Neher, P., Götz, M., Norajitra, T., Weber, C., & Maier-Hein, K. (2015). A Machine Learning Based Approach to Fiber Tractography Using Classifier Voting. In Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015 (pp. 45–52). Springer International Publishing.
- Neher, P., Côté, M.A., Houde, J.C., Descoteaux, M., & Maier-Hein, K. (2017). Fiber tractography using machine learning. NeuroImage, 158, 417–429
- Benou, I., & Raviv, T. (2019). Deeptract: A probabilistic deep learning framework for white matter fiber tractography. In International Conference on Medical Image Computing and Computer-Assisted Intervention (pp. 626–635).
- Viktor Wegmayr, & Joachim M. Buhmann (2020). Entrack: Probabilistic Spherical Regression with Entropy Regularization for Fiber Tractography. International Journal of Computer Vision.
- Wegmayr, V., Giuliari, G., Holdener, S., & Buhmann, J. (2018). Data-driven fiber tractography with neural networks. In 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018) (pp. 1030–1033).
Figures
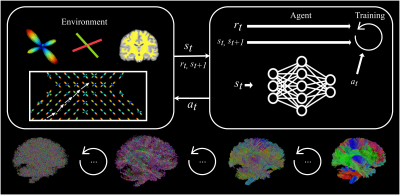
Representation of the framework. Top: The RL loop, where states, rewards and actions are exchanged between the learning agent and the environment. Top-left: the environment keeps track of the reconstructed streamlines and computes states and rewards accordingly. Top-right: The agent uses states and rewards received to improve itself and output actions. Bottom: Reconstructed tractograms are iteratively more plausible as training goes on.
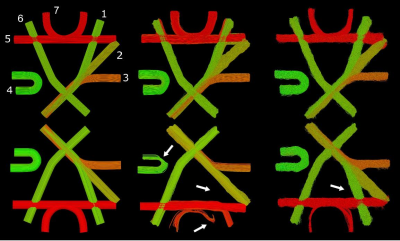
Reconstructions of bundles of both FiberCups8,9. Top: Reconstructions of the original FiberCup. Bottom: Reconstruction of the “flipped” FiberCup. Left: Ground-truth bundles, numbered for clarity. Center: Reconstruction by Learn-to-Track10. Right: Reconstruction by our SAC agent. Indicated by arrows, we can see in the center column that Learn-to-Track failed to reconstruct bundle 1, and that bundles 4 and 7 are poorly reconstructed, whereas our reconstructions are consistent.
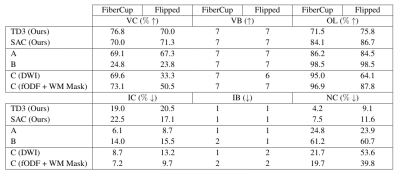
Results for experiment 1. Valid Connections (VC), Valid Bundles (VB), Overlap (OL), Invalid Connections (IC), Invalid Bundles (IB) and No-Connections (NC) are reported by the Tractometer13. A: classical deterministic algorithm with fODFs input; B: classical probabilistic algorithm with same input; C: Learn-toTrack9, with either raw diffusion as input (DWI), or the same input as the proposed method (fODF + WM Mask).
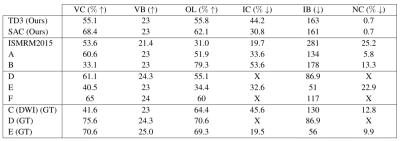
Results for experiment 2. A, B, C refer to the same methods as in Figure 3. D refers to Neher et al.15,16, E refers to Benou et al.17, F refers to Wegmayr et al. (2020)18. ISMRM2015 refers to the mean results of the original challenge11. (GT) indicates that the method was trained on the ground-truth bundles. X indicates measures that were not reported by the original authors. Reported metrics are the same as in Figure 3.
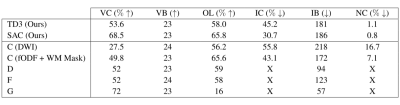
Results for experiment 3. A, B, C, D, F refer to the same methods as in Figure 3. G refers to Wegmayr et al. (2018)19 . X indicates measures that were not reported by the original authors. Reported metrics are the same as in Figure 3.