0329
Fast and Accurate Modeling of Transient-state Sequences by Recurrent Neural Networks1Computational Imaging Group for MR diagnostics & therapy, Center for Image Sciences, UMC Utrecht, Utrecht, Netherlands
Synopsis
Fast and accurate modeling of transient-state sequences are required for various quantitative MR applications. We present here a surrogate model based on Recurrent Neural Network (RNN) architecture, to quickly compute large-scale MR signals and derivatives. We demonstrate that the trained RNN model works with different sequence parameters and tiussue parameters without the need of retraining. We prove that the RNN model can be used for computing large-scale MR signals and derivatives within seconds, and therefore achieves one to three orders of magnitude acceleration for different qMRI applications.
Introduction
Fast and accurate modeling of MR signal responses are typically required for quantitative MRI applications, such as MR Fingerprinting (MRF)1 and MR-STAT2. Taking MR Fingerprinting dictionary generation as an example, the computational time for simulating large amount of MR signals can be prohibitively long, from hours to days, especially when slice profile effects are taken into account3.Based on recent development in deep learning, we propose a Recurrent Neural Network (RNN) architecture with multiple stacked layers for quickly computing large-scale MR signals and derivatives for transient-state gradient-spoiled sequences. Two main advantages of the RNN model are its generalization capability and computational efficiency: the same RNN model works with different sequence parameters, such as sequence length and time-dependent flip-angle train without need for retraining. We show that the RNN surrogate model can be used for orders of magnitude acceleration of different qMRI applications, in particular MRF dictionary generation and optimal experimental design.
Methods
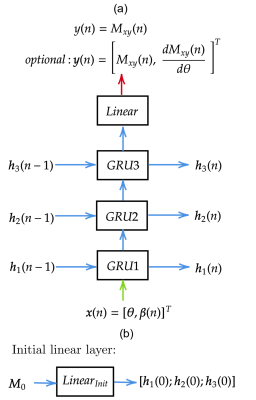
- Network architecture
- Data generation and network training
The dataset was split into a training set of size 20000 and a test set of size 10000. The RNN network was built and trained using Tensorflow 2.2 on a Tesla-V100-GPU, run for 3000 epochs, ADAM optimizer using Mean Absolute Error loss function, batch size 200.
- Applications
The second example is MRF sequence optimization. Specifically, the problem is to find the optimal flip-angle train for minimizing the reconstruction noise which is characterized by the Cramér-Rao lower bound (CRLB)8. We conducted a simulation-based experiment to optimize a Spline11-type flip-angle train given two target tissues with $$$T_1/T_2=900/85$$$ms and $$$T_1/T_2=500/65$$$ms. The sequence length is 336 with $$$T_E/T_R=4.9/8.7$$$ms. Constrained differential evolution was used for solving the optimization problem, and the RNN model was used for accelerating the CLRB computation.
Results
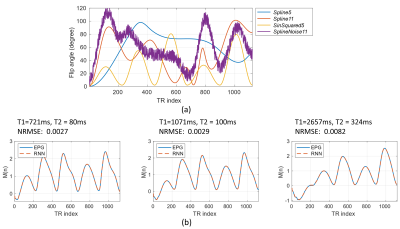
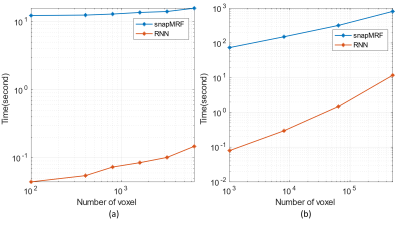
The total training took approximately 8 hours. Fig.2(b) shows sample RNN signals are in excellent agreement with EPG signals. The overall NRMSEs for test dataset are relatively low: 0.77% for magnetization signals and 1.39% for signal derivatives, showing that the proposed RNN is able to accurately model the signal even for unseen flip angle trains.Fig.3 shows the runtime comparison results for our RNN and the recently proposed fast GPU simulator snapMRF9. In Fig.3(a) and 3(b), all the runtime curves grow approximately linearly with respect to the number of signals. For the fixed $$$B_1^+$$$ condition in Fig.3(a), RNN requires ~100 times shorter runtime than snapMRF for a dataset with 6400 signals. For various $$$B_1^+$$$ conditions (a large dataset with 512000 signals) , our RNN outperforms snapMRF by a factor of 68 (Fig.3(b)).
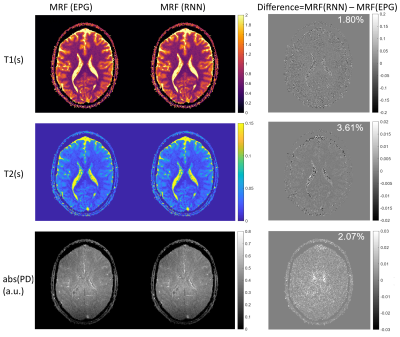
Fig.4 shows that in-vivo MRF reconstructions using EPG or RNN generated dictionaries are very similar. Generating the dictionary with 7812 signals by RNN takes just 0.3s, whereas running the EPG model on CPU takes about 1.7 hours.
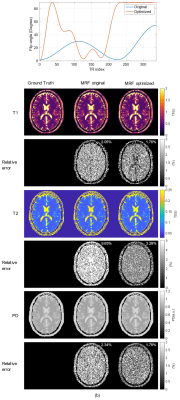
Fig.5(a) shows the optimized flip-angle train and Fig.5(b) shows the MRF-reconstructed $$$T_1, T_2$$$ and $$$abs(PD)$$$ maps using the original and optimized sequences. The optimized sequence improves the accuracy of all the three reconstructed maps comparing to the original sequence, with a most significant improvement in $$$T_2$$$ maps. Solving the sequence optimization problem now takes about 10.5 seconds, whereas in previous works10, solving similar problems requires at least one CPU hour.
Discussion and Conclusion
This work proposed a RNN model as a fast surrogate of the EPG model for computing large-scale MR signals and derivatives. We demonstrated that the RNN model is between one and three orders of magnitude faster than the GPU-accelerated EPG package snapMRF9. The RNN surrogate model can be efficiently used for computing large-scale MRF dictionary signals and derivatives within seconds. The practical application of transient-state quantitative techniques can therefore be substantially facilitated. In the future, usage of the RNN model may be extended for other types of sequences, or for modeling more complex physics such as magnetic transfer effects. Code is freely available at https://gitlab.com/HannaLiu/rnn_epg.Acknowledgements
The first author receives CSC(Chinese Scholarship Counsel) scholarship.References
[1] Ma D, Gulani V, Seiberlich N, et al. Magnetic resonance fingerprinting. Nature. 2013;495(7440):187-192.
[2] Sbrizzi A, van der Heide O, Cloos M, et al. Fast quantitative MRI as a nonlinear tomography problem. Magn Reson Imaging. 2018;46:56-63.
[3] Ostenson J, Smith DS, Does MD, Damon BM. Slice-selective extended phase graphs in gradient-crushed, transient-state free precession sequences: An application to MR fingerprinting. Magn Reson Med. 2020.
[4] Hermans M, Schrauwen B. Training and analysing deep recurrent neural networks. In: Advances in Neural Information Processing Systems. ; 2013:190-198.
[5] Liu H, van der Heide O, van den Berg CAT, Sbrizzi A. Fast and Accurate Modeling of Transient-state Gradient-Spoiled Sequences by Recurrent Neural Networks. arXiv Prepr arXiv200807440. 2020.
[6] van der Heide O, Sbrizzi A, Bruijnen T, van den Berg CAT. Extension of MR-STAT to non-Cartesian and gradient-spoiled sequences. In: Proceedings of the 2020 Virtual Meeting of the ISMRM. 2020:0886.
[7] Assländer J, Cloos MA, Knoll F, Sodickson DK, Hennig J, Lattanzi R. Low rank alternating direction method of multipliers reconstruction for MR fingerprinting. Magn Reson Med. 2018;79(1):83-96.
[8] Zhao B, Haldar JP, Liao C, et al. Optimal experiment design for magnetic resonance fingerprinting: Cramer-Rao bound meets spin dynamics. IEEE Trans Med Imaging. 2018;38(3):844-861.
[9] Wang D, Ostenson J, Smith DS. snapMRF: GPU-accelerated magnetic resonance fingerprinting dictionary generation and matching using extended phase graphs. Magn Reson Imaging. 2020;66:248-256.
[10] Lee PK, Watkins LE, Anderson TI,
Buonincontri G, Hargreaves BA. Flexible and efficient optimization of
quantitative sequences using automatic differentiation of Bloch simulations. Magn Reson Med. 2019;82(4):1438-1451.
Figures