0290
NIfTI MRS: A standard format for spectroscopic data1Wellcome Centre for Integrative Neuroimaging, NDCN, University of Oxford, Oxford, United Kingdom, 2Department of radiology, University of Calgary, Calgary, AB, Canada, 3Hotchkiss brain institute, University of Calgary, Calgary, AB, Canada, 4Alberta Children’s Hospital Research Institute, University of Calgary, Calgary, AB, Canada, 5School of Health Sciences, Purdue University, West Lafayette, IN, United States, 6Weldon School of Biomedical Engineering, Purdue University, West Lafayette, IN, United States, 7Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 8F. M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, Baltimore, MD, United States, 9Division of Cancer Science, School of Medical Sciences, Faculty of Biology, Medicine and Health, University of Manchester, Manchester, United Kingdom, 10Institute of Scientific Instruments of the CAS, Brno, Czech Republic, 11Department of Biomedical Engineering, Faculty of Electrical Engineering and Communication, Brno University of Technology, Brno, Czech Republic, 12Center for Advanced MR Development, Department of Radiology, Duke University Medical Center, Durham, NC, United States, 13Centre for Human Brain Health and School of Psychology, University of Birmingham, Birmingham, United Kingdom
Synopsis
We propose a flexible data format for the storage of multidimensional MRS data. The lack of a single widely used data format in MRS hinders widespread use, consistent analyses, and open dissemination of data. Here, we adapt the widely adopted NIfTI format to store MRS data, extending NIfTI to include the additional meta-data required for MRS data interpretation. We present a pipeline for end-to-end analysis of MRS data and an open-source conversion tool. The community is encouraged to contribute to the format.
Introduction
Users of magnetic resonance spectroscopy (MRS) and spectroscopic imaging (MRSI) are faced with numerous data formats from the scanner, both proprietary and DICOM.1 The DICOM standard for MRS is not fully or consistently implemented by the vendors, and crucial information is stored in private tags. Therefore, vendor-specific expertise is needed to interpret, reconstruct, and analyse MRS data. Formats are frequently modified and not typically documented, and interpretation relies on disparate software, with inconsistent implementations.Furthermore, there is no accepted format for sharing or storing spectral data. Researchers cannot easily interpret data processed in another laboratory, nor can data be made “open” in a consistent format. A single MRS(I)-specific format would solve these problems, as well as paving the way for modular analysis: promoting validated discrete analysis steps.
We propose using an extended Neuroimaging Informatics Technology Initiative (NIfTI) format to store MRS data, referred hereafter as "NIfTI MRS".2,3 NIfTI has been chosen due to its simplicity, availability of supporting libraries, ubiquity in the neuroimaging community, and extensive pre-existing support. Moreover, the results of spectral fitting can trivially be stored as unmodified NIfTI images.
By implementing the single format we aim to:
- Provide a simplified pathway from scanner to final analysis,
- Enable interoperability and modularity of analysis programs,
- Enable easier display and co-interpretation with other modalities, and
- Establish a format for data sharing.
We envisage the adoption of NIfTI MRS will make MRS accessible to a wider group of researchers, who will be able to run standardized and automated data analysis and more easily share their data - ultimately improving the reproducibility and transparency of MRS research.
Methods
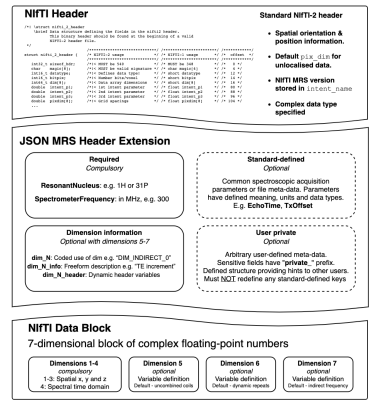
NIfTI MRS uses the base NIfTI format to contain the complex time-domain data, spatial orientation information, and the spectral dwell time. Additional JavaScript Object Notation (JSON)-formatted metadata stores information essential for interpretation as a NIfTI header extension; for minimum conformance, storing the spectrometer frequency and the resonant nucleus is required.4,5The NIfTI MRS data array has seven dimensions. The first four mandatory dimensions correspond to spatial x, y and z directions, and time. The use of the last three optional dimensions is flexible and specified using the header extension, with coil elements, dynamic repeats, and indirect frequency (for 2D experiments) assigned as defaults.
NIfTI MRS rigidly defines further optional header extension fields. These fields define common acquisition parameters used in MRS analysis and scanner, sequence, and subject information. Furthermore, users may define their own fields, conforming to a fixed structure, to allow flexible adaptation to future sequence and software development. Simplified de-identification is achieved using a predefined list of “at-risk” fields and explicit notation for user-defined fields.
Each element of the three optional data dimensions can be associated with different meta-data values, e.g., the unique acquisition conditions of each transient of a “fingerprinting” experiment can be stored.
Results
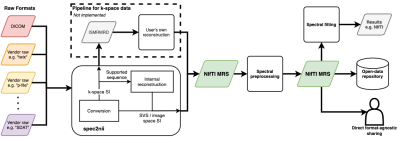
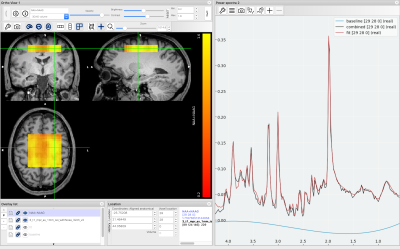
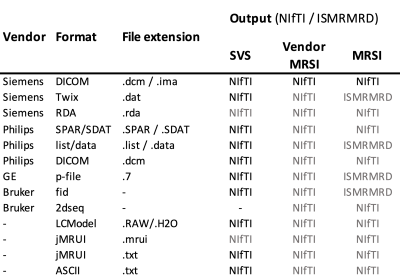
Figure 1 shows a schematic of the proposed NIfTI MRS format, with the full specification available online at Reference 6 alongside example data. The specification was developed and agreed by the authors and forms the core of a proposed analysis workflow - illustrated in Figure 2. Data converted to the NIfTI MRS format is shown, as displayed in FSLeyes, in Figure 3.7 To provide this conversion a command-line conversion program spec2nii has been developed, currently operating on a limited set of data formats. Figure 4 gives a summary of known and converted formats. Finally, Figure 5 highlights packages committed to supporting this format and shows SVS data in NIfTI MRS loaded in three of those packages.Discussion & Conclusion
We have developed a new standard for the storage of MRS data, based on the ubiquitous NIfTI file format. Eight MRS analysis packages intend to support the format (Figure 5) and we further encourage adoption by providing example input-output code at Reference (6).A single point of conversion from vendor formats to NIfTI MRS is being developed (github.com/wexeee/spec2nii). We encourage MRS researchers to suggest improvements and submit example data to aid community development of robust conversion tools. In the future, spatial reconstruction of vendor MRSI sequences will be implemented to ease access of NIfTI MRS. Because NIfTI MRS cannot store arbitrary k-space data, nor reconstruct arbitrary data automatically, fallback conversion to ISMRM Raw Data (ISMRMRD) format will be provided.8,9
Neuroimaging has benefited from widespread adoption of the NIfTI format, with open-source analysis tools (AFNI, FSL, SPM) providing native support.10–12 By extending NIfTI to store MRS data we aim to harmonise the storage, processing, and multi-modal visualisation of MRS with MRI. The development of NIfTI MRS greatly simplifies the integration of MRS into existing neuroimaging initiatives - such as BIDS and openneuro.13,14
We have released an initial draft of a new format for MRS data which will: 1) provide a common format for data sharing; 2) enable interoperability between MRS analysis software; 3) reduce the burden of supporting vendor formats by encouraging an open community-based support model and 4) harmonise MRS data storage with other modalities to leverage existing software, initiatives and data sharing infrastructure. To ensure suitability for the wide range of MRS techniques the authors encourage and request feedback from the community (at tinyurl.com/yd6sxb2s and forum.mrshub.org).
Acknowledgements
The Wellcome Centre for Integrative Neuroimaging is supported by core funding from the Wellcome Trust (203139/Z/16/Z).References
1. The DICOM standard. Section C.8.14 MR Spectroscopy Modules. http://dicom.nema.org/medical/dicom/2017c/output/chtml/part03/sect_C.8.14.html. Accessed December 16, 2020.
2. NIfTI-2 Data Format — Neuroimaging Informatics Technology Initiative. https://nifti.nimh.nih.gov/nifti-2. Accessed December 16, 2020.3. NIfTI Documentation. https://nifti.nimh.nih.gov/pub/dist/src/niftilib/nifti1.h. Accessed December 16, 2020.
4. Pezoa F, Reutter JL, Suarez F, Ugarte M, Vrgoč D. Foundations of JSON schema. In: Proceedings of the 25th International Conference on World Wide Web. International World Wide Web Conferences Steering Committee; 2016. pp. 263–273.
5. JSON. https://www.json.org/json-en.html. Accessed December 16, 2020.
6. Clarke WT. github.com/wexeee/mrs_nifti_standard. Accessed December 16, 2020.
7. McCarthy P. FSLeyes. Zenodo; 2020. doi: 10.5281/zenodo.3937147.
8. Inati SJ, Naegele JD, Zwart NR, et al. ISMRM Raw data format: A proposed standard for MRI raw datasets. Magn. Reson. Med. 2017;77:411–421 doi: https://doi.org/10.1002/mrm.26089.
9. ISMRM Raw Data Format (ISMRMRD) — ISMRMRD 1.1.0 documentation. https://ismrmrd.github.io/. Accessed December 16, 2020.
10. Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM. FSL. NeuroImage 2012;62:782–790 doi: 10.1016/j.neuroimage.2011.09.015.
11. Friston KJ ed. Statistical parametric mapping: the analysis of funtional brain images. 1st ed. Amsterdam ; Boston: Elsevier/Academic Press; 2007.
12. Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. Int. J. 1996;29:162–173 doi: 10.1006/cbmr.1996.0014.
13. Poldrack RA, Gorgolewski KJ. OpenfMRI: Open sharing of task fMRI data. NeuroImage 2017;144:259–261 doi: 10.1016/j.neuroimage.2015.05.073.
14. Gorgolewski KJ, Auer T, Calhoun VD, et al. The brain imaging data structure, a format for organizing and describing outputs of neuroimaging experiments. Sci. Data 2016;3:160044 doi: 10.1038/sdata.2016.44.
Figures