0196
VAE deep learning model with domain adaptation and harmonization for diagnostic classification from multi-site neuroimaging data1Electrical and Computer Engineering, AU MRI Research Center, Auburn University, Auburn, AL, United States, 2Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Medical School and Harvard-MIT Health Sciences and Technology, Charlestown, MA, United States, 3Symbiosis Center for Medical Image Analysis, Symbiosis International University, Pune, India
Synopsis
In large public multi-site fMRI datasets, the sample characteristics, data acquisition methods and pre-processing approaches vary across sites and datasets, leading to poor diagnostic classification. Domain adaptation aims to improve the classification performance in target domain data by utilizing the knowledge learned from the source domain, and making the distributions of data in source and target domains as similar as possible. In this sense, domain adaptation is one method that can be used to achieve and optimize transfer learning by using different datasets.
Introduction
Deep-learning models outperform traditional machine learning methods in identifying individuals with psychiatric disorders1-3, including Autism4-6. Large public databases such as ABIDE (Autism Brain Imaging Data Exchange) have aided deep-learning models in this endeavor. However such large public databases have been assembled post-hoc, and hence contain different sources of non-neural variability such as different sites using different scanners and protocols, which degrade performance of deep-learning7,8. To address this, we propose domain-adaptation, which aims to improve the classification performance in a target-domain by utilizing the knowledge learned from the source-domain by making the distributions of data in source and target-domains as similar as possible9,10. This is in contrast to statistical ComBat-harmonization, which relies on statistical modeling as opposed to deep-learning used by domain-adaptation, to achieve the same goal11-13. However, for classification using deep-learning, domain-adaptation provides the advantage that it uses the same deep-learning concepts used in classification, and hence can be elegantly integrated within the same framework. It is also expected to work better with increasing data size.Therefore, we developed a domain-adaptation framework using variational auto-encoder (VAE) with maximum-mean-discrepancy (MMD) regularization, to minimize the inter-site variance across ABIDE-I and ABIDE-II datasets. We utilized both deep-learning and ComBat-harmonization approaches to evaluate their efficacy when used in isolation or in combination. To test the hypothesis that accuracy obtained from domain-adaptation in multisite data can be improved further with inclusion of more data, we included healthy-control data from large-scale datasets such as Amsterdam Open MRI Collection (AOMIC)14 and Health Brain Network (HBN)15 into the source-domain.Methods
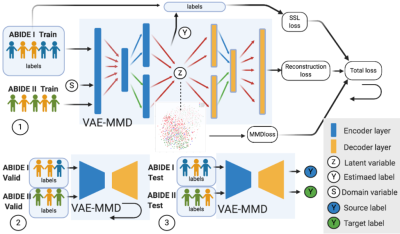
We propose a VAE-MMD16-19 model for applying domain-adaptation to ASD classification. In feature extraction and training process, MMD regularization term and VAE were jointly optimized. The framework is illustrated in Fig.1. Both encoder and decoder networks have two layers. The first layer is constructed as the latent-feature discriminative model (M1) and the second layer is constructed as a generative semi-supervised model (M2) in stacked architecture20. The learning rate is equal to 0.0005. Each NN layer uses ReLU as activation function. Drop-out layer is also applied in training, the drop-out rate being 0.821.In the 3-way (health control, Asperger’s and autism) ASD classification task, we collected ABIDE-I (n=988) as the labeled supervised dataset and ABIDE-II (n=623) as the unlabeled unsupervised dataset. We set ABIDE-I as source-domain, and ABIDE-II as target-domain. These data are split into three sets uniformly sampled across sites: training, validation and test datasets. Additional healthy-control datasets from AOMIC (n=334) and HBN (n=337) were subsequently included into the source-domain to check whether there is scope for further improvement in accuracy with inclusion of more data. Pre-processing and estimation of functional-connectivity matrices, which were used as input features to the classifiers, were performed by CONN toolbox 22.During training we input both ABIDE-I with labels and unlabeled ABIDE-II training datasets into VAE-MMD model. ABIDE-I and ABIDE-II validation datasets were used in validation process to fine-tune the hyperparameters α and β. The VAE-MMD approach learns the invariant representations across different domains while retaining the discriminative information required for classification. For comparison, we also use a baseline approach without domain-adaptation wherein we set as zero so that no MMD regularization is carried out in optimization. We visualized the process of domain-adaptation using t-distributed stochastic neighbor embedding (t-SNE) plots23.Results
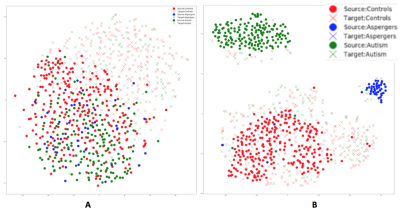
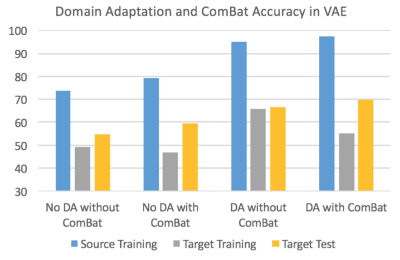
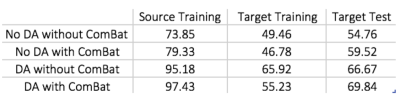
Fig.2 visualizes the feature space before and after domain-adaptation using t-SNE23 plots, showing how domain-adaptation helps separate the feature space belonging to different groups in multi-site data. Table.1/Fig.3 illustrates that both ComBat and domain-adaptation can improve classification, especially when used in combination. For three-way classification using test-data in the target-domain, our model achieved 69.84% accuracy with domain-adaptation and ComBat. Given the smaller number of samples from Asperger’s and its similarities with autism, three-way classification in ABIDE is a hard problem. Although cross-validation accuracies above chance (which is 33%) have been reported before, accuracy in independent test data rarely exceeded 50%8. If we included AOMIC and HBN datasets into the source-domain, accuracy further increased to 76.19%, demonstrating that there is scope within the domain-adaptation framework to improve the accuracy further by including more data.Discussion
Even with high-dimensional input-features, the VAE-MMD model outperforms by projecting data-points from different domains from the same class into a closed latent-space. Our results demonstrate that deep-learning based domain-adaptation and statistical ComBat-harmonization approaches can improve the target-domain classification when used independently. When used in combination, the accuracy seems to be better than when they are used independently, indicating that both methods bring something unique to the table. Specifically, Fig.3/Table.1 show that learning from labeled training data in the source-domain improves dramatically with domain-adaptation and ComBat-harmonization, with same trends seen in the target-domain with unlabeled data, but to a lesser extent24. t-SNE visualization demonstrates how domain-adaptation separates features obtained from multi-site data in the feature space. In the absence of such inter-site harmonization efforts, the inter-class differences are drowned amidst the large inter-site variability.Conclusion
We applied deep-learning-based domain-adaptation and statistical ComBat-harmonization for improving diagnostic accuracy from multi-site neuroimaging data and show robust performance of our approach in the context of diagnostic classification of ASD.Acknowledgements
No acknowledgement found.References
1. Durstewitz, Daniel & Koppe, Georgia & Meyer-Lindenberg, Andreas. (2019). Deep neural networks in psychiatry. Molecular Psychiatry. 24. 1. 10.1038/s41380-019-0365-9.
2. Wen, D., Wei, Z., Zhou, Y., Li, G., Zhang, X., & Han, W. (2018). Deep Learning Methods to Process fMRI Data and Their Application in the Diagnosis of Cognitive Impairment: A Brief Overview and Our Opinion. Frontiers in neuroinformatics, 12, 23. doi:10.3389/fninf.2018.00023
3. Vieira, S., Pinaya, W.H., & Mechelli, A. (2017). Using deep learning to investigate theneuroimaging correlates of psychiatric and neurological disorders: Methods and applications. Neuroscience & Biobehavioral Reviews, 74, 58-75.
4. Nielsen, J.A., et al., (2013). Multisite functional connectivity MRI classification of autism: ABIDE results. Front. Hum. Neurosci. 7 (September), 1–12.
5. Di Martino, A., O’Connor, D., Chen, B. et al. Enhancing studies of the connectome in autism using the autism brain imaging data exchange II. Sci Data 4, 170010 (2017)
6. Horien, C., Noble, S., Greene, A.S. et al. A hitchhiker’s guide to working with large, open-source neuroimaging datasets. Nat Hum Behav (2020).
7. Nielsen, J. A., Zielinski, B. A., Fletcher, P. T., Alexander, A. L., Lange, N., Bigler, E. D., Lainhart, J. E., & Anderson, J. S. (2013). Multisite functional connectivity MRI classification of autism: ABIDE results. Frontiers in human neuroscience, 7, 599.
8. P Lanka, D Rangaprakash, MN Dretsch, JS Katz, TS Denney, G Deshpande, "Supervised machine learning for diagnostic classification from large-scale neuroimaging datasets", Brain Imaging and Behavior, 2019 (in press).
9. X. Li, Y. Gu, N. Dvornek, L. Staib, P. Ventola and J. Duncan, "Multi-site fMRI Analysis Using Privacy-preserving Federated Learning and Domain Adaptation: ABIDE Results," Medical Image Analysis, p. 101765, 2020.
10. Zhou, S., Cox, C., & Lu, H. (2018). Improving Whole-Brain Neural Decoding of fMRI with Domain Adaptation. bioRxiv.
11. Jean-Philippe Fortin, Drew Parker, Birkan Tunc, Takanori Watanabe, Mark A Elliott, Kosha Ruparel, David R Roalf, Theodore D Satterthwaite, Ruben C Gur, Raquel E Gur, Robert T Schultz, Ragini Verma, Russell T Shinohara. Harmonization Of Multi-Site Diffusion Tensor Imaging Data. NeuroImage, 161, 149-170, 2017
12. Fortin, J. P., Cullen, N., Sheline, Y. I., Taylor, W. D., Aselcioglu, I., Cook, P. A., Adams, P., Cooper, C., Fava, M., McGrath, P. J., McInnis, M., Phillips, M. L., Trivedi, M. H., Weissman, M. M., & Shinohara, R. T. (2018). Harmonization of cortical thickness measurements across scanners and sites. NeuroImage, 167, 104–120.
13. W. Evan Johnson and Cheng Li, Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics, 8(1):118-127, 2007.
14. Snoek LvdM, M.M; Beemsterboer, T.; van der Leij, A.; Eigenhuis, A.; Scholte, H.S. The Amsterdam Open MRI Collection, a set of multimodal MRI datasets for individual difference analyses. bioRxiv, (2020).
15. Alexander LM, et al. An open resource for transdiagnostic research in pediatric mental health and learning disorders. Sci Data 4, 170181 (2017).
16. D. Kingma and M. Welling, "Auto-Encoding Variational Bayes," 2014.
17. M. Belhaj, P. Protopapas and W. Pan, "Deep Variational Transfer: Transfer Learning through Semi-supervised Deep Generative Models.," 2018.
18. H. Chen and J. Chien, "Deep semi-supervised learning for domain adaptation," in 2015 IEEE 25th International Workshop on Machine Learning for Signal Processing (MLSP), Boston, MA, 2015.
19. A. Rahimi and B. Recht, "Weighted sums of random kitchen sinks: Replacing minimization with randomization in learning," In Advances in neural information processing systems, p. 1313– 1320, 2009.
20. C. Louizos, K. Swersky, Y. Li, M. Welling and R. Zemel, "The variational fair autoencoder," in International Conference on Learning Representations (ICLR), 2016.
21. Srivastava, Nitish & Hinton, Geoffrey & Krizhevsky, Alex & Sutskever, Ilya & Salakhutdinov, Ruslan. (2014). Dropout: A Simple Way to Prevent Neural Networks from Overfitting. Journal of Machine Learning Research. 15. 1929-1958.
22. Whitfield-Gabrieli, S., & Nieto-Castanon, A. (2012). Conn: a functional connectivity toolbox for correlated and anticorrelated brain networks. Brain connectivity, 2(3), 125-141
23. L. v. d. Maaten and G. Hinton, "Visualizing data using t-sne," Journal of machine learning research, vol. 9, no. Nov, p. 2579–2605, 2008.
24. W. M. Kouw and M. Loog, "A review of domain adaptation without target labels," in IEEE Transactions on Pattern Analysis and Machine Intelligence, doi: 10.1109/TPAMI.2019.2945942.
Figures