0187
SIMBA 2.0: An enhanced SImilarity-driven Multi-dimensional Binning Algorithm for free-running ferumoxytol-enhanced whole-heart MRI1Advanced Clinical Imaging Technology, Siemens Healthcare AG, Lausanne, Switzerland, 2Department of Radiology, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 3IHU LIRYC, Electrophysiology and Heart Modeling Institute, Fondation Bordeaux Université, Pessac-Bordeaux, France, 4Department of Cardiovascular Imaging, Hôpital Cardiologique du Haut-Lévêque, CHU de Bordeaux, Pessac, France, 5Division of Pediatric Cardiology, Department Woman-Mother-Child, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 6Division of Cardiology, Cardiovascular Department, Lausanne University Hospital and University of Lausanne, Lausanne, Switzerland, 7CIBM Center for Biomedical Imaging, Lausanne, Switzerland, 8Cardiac MR Center, Lausanne University Hospital, Lausanne, Switzerland
Synopsis
A SImilarity-driven Multi-dimensional Binning Algorithm (SIMBA) was recently proposed for fast reconstruction of motion-consistent clusters for free-running whole-heart MRA acquisitions. Originally, only the most populated cluster was used for the reconstruction of a motion-suppressed image. In this work we investigated whether the redundancy of information among the clusters can be exploited to improve image quality. Specifically, an adapted XD-GRASP reconstruction and a multidimensional patch-based low-rank denoising algorithm were compared. Four different reconstructions were quantitatively evaluated and compared using ferumoxytol-enhanced free-running datasets from 10 pediatric and adult CHD patients. Information sharing resulted in significantly sharper anatomical features and increased image quality.
Background
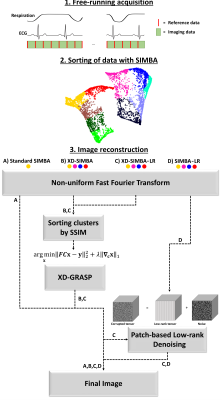
As recently reported1, a novel approach to physiological motion compensation was proposed and applied to free-running whole-heart MRI datasets2. This SImilarity-driven Multi-dimensional Binning Algorithm (SIMBA) provides a fast (~1 min) reconstruction for static motion-suppressed datasets without explicit physiological signal extraction and without any assumptions regarding cardiac and respiratory frequencies. Briefly, SIMBA exploits the inherent similarities in the acquired data by projecting the instances of a repeated reference readout as data points into an n-dimensional space and subsequently binning such points into disjoint clusters. It was demonstrated that each cluster intrinsically provides a static sub-image from a specific respiratory and cardiac motion state. In1, only the most populated cluster was reconstructed and analyzed as a proof of principle. This approach can be considered analogous to a retrospective cardiac and respiratory gating, where only data from a specific physiological state are used for reconstruction, while the rest is discarded. As mentioned above, however, it was shown that all SIMBA clusters produce static sub-images from different physiological states and, therefore, there is a richness of redundant information about cardiac structure that can potentially be exploited for enhanced multi-cluster image reconstruction. The XD-GRASP approach3, for instance, proposes to use the correlation between similar motion-states to produce images of improved quality through a k-t sparse SENSE iterative reconstruction4,5. Here, a reconstruction obtained by applying the XD-GRASP concept to multiple SIMBA clusters is compared to the single cluster reconstruction. Additionally, local similarities both within and among the different clusters are also exploited by using a modified version of the multi-dimensional patch-based low-rank denoising method proposed by Bustin et al.6 after reconstruction.Materials and Methods
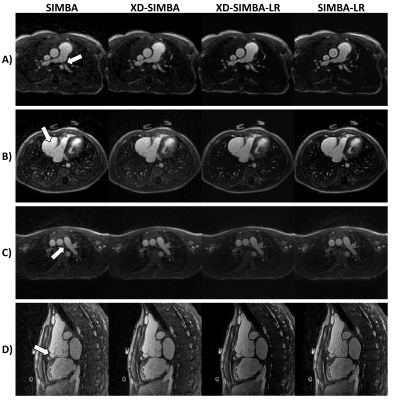
Ten congenital heart disease patients, including both children and adults (age: 23±20 years, weight: 58±34 kg, 7 Males), with clinical indication for cardiac MRI, were included in this IRB approved study. Examinations were performed during free-breathing, either un-sedated or with light oral sedation, on a 1.5T clinical MRI system (MAGNETOM Sola, Siemens Healthcare, Erlangen, Germany) after administration of 2 mg/kg of ferumoxytol7. A slab-selective spoiled gradient echo adaptation of the prototype free-running 3D radial acquisition strategy described in2 was used and resulted in uninterrupted acquisitions of 5:35–5:59 minutes duration. Main sequence parameters were: RF excitation angle: 15°, resolution: (1.15-1.35 mm)3, FOV (220-260 mm)3, TE/TR: 1.53-1.64/2.69-2.84 ms, readout bandwidth: 1002 Hz/pixel.Image reconstruction: Each of the free-running datasets was reconstructed with four different techniques: 1) the original single-cluster version of SIMBA1 as a reference standard, 2) XD-GRASP reconstruction incorporating the four most populated SIMBA clusters (XD-SIMBA), 3) XD-SIMBA followed by the patch-based low-rank denoising approach (XD-SIMBA-LR), and 4) the denoising approach directly applied to the reconstructions of the four most populated SIMBA clusters (SIMBA-LR) (Figure 1). Before applying the XD-GRASP reconstruction to exploit the correlation along the cluster dimension, the structural similarity index measure (SSIM)8 was used to sort the clusters in terms of similarity. The multi-dimensional patch-based low-rank denoising method was used here as a one-step denoising, as opposed to the iterative scheme proposed in6. The denoising procedure was performed by exploiting local similarities both slice-by-slice and along the cluster dimension, in axial, coronal and sagittal directions.
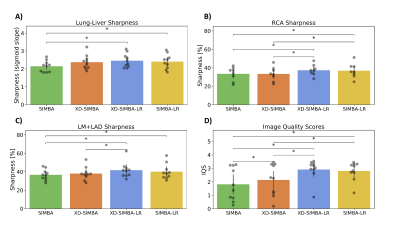
Data Analysis: After a visual comparison of the general image quality, the sharpness of the lung-liver interface was quantified similarly to9. The sharpness of the right coronary artery (RCA) and the left main (LM) + left anterior descending (LAD) coronary arteries were compared using Soap-Bubble10 on multiplanar reformats. Finally, a recently published convolutional neural network was used for automated image quality assessment11.
Results
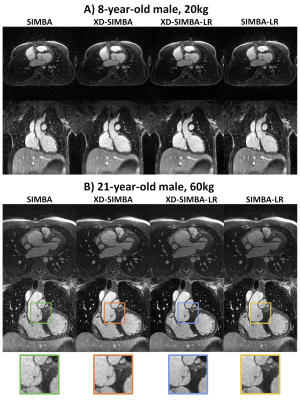
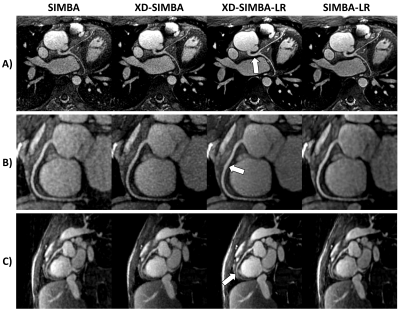
Visually, the strategies that exploit information redundancy between clusters resulted in sharper and less noisy images (Figure 2). XD-SIMBA, XD-SIMBA-LR, and SIMBA-LR provided improved lung-liver interface sharpness when compared to standard SIMBA (Figure 3A). The multiplanar reformats in XD-SIMBA-LR appear to be less noisy and sharper than those from the other reconstruction techniques (Figure 4). This was corroborated by the vessel sharpness, as XD-SIMBA-LR images on average had the sharpest LM+LADs and RCAs (Figure 3C-3B). The scores from the convolutional neural network are shown in Figure 3D. XD-SIMBA-LR scored significantly higher than SIMBA and XD-SIMBA (P<0.05), but not significantly higher than SIMBA-LR (P=0.12). Examples of anatomical anomalies are provided in Figure 5.Discussion and Conclusions
This work focused on exploiting the redundant information in the SIMBA clusters to reconstruct free-running ferumoxytol-enhanced whole-heart MRI. Although XD-SIMBA, XD-SIMBA-LR, and SIMBA-LR in general provided improved image quality when compared to SIMBA alone, the effect sizes were relatively modest. To further increase the benefit from the redundancy of information, the clusters could be co-registered prior to or within the reconstruction procedures. The denoising was applied as a one-step strategy whereas the total variation constraint in the XD-GRASP reconstructions was part of an iterative reconstruction scheme. Integration of the denoising as a regularization factor in an iterative approach6 will certainly be considered moving forward. In conclusion, it has been demonstrated that the richness of redundant information among the SIMBA clusters can be exploited by using the low-rank properties of similar image patches and/or the global similarity as represented by finite differences between cluster images. This SIMBA2.0 approach produces improved image quality and coronary sharpness with respect to the original single-cluster SIMBA approach.Acknowledgements
No acknowledgement found.References
[1] J. Heerfordt et al., “Free-running SIMilarity-Based Angiography (SIMBA) for simplified anatomical MR imaging of the heart” arXiv preprint arXiv:2007.06544
[2] L. Di Sopra, D. Piccini, S. Coppo, M. Stuber, and J. Yerly, “An automated approach to fully self‐gated free‐running cardiac and respiratory motion‐resolved 5D whole‐heart MRI,” Magn. Reson. Med., vol. 82, no. 6, pp. 2118–2132, Dec. 2019.
[3] L. Feng, L. Axel, H. Chandarana, K. T. Block, D. K. Sodickson, and R. Otazo, “XD-GRASP: Golden-angle radial MRI with reconstruction of extra motion-state dimensions using compressed sensing,” Magn. Reson. Med., vol. 75, no. 2, pp. 775–788, 2016.
[4] D. Piccini et al., “Four-dimensional respiratory motion-resolved whole heart coronary MR angiography,” Magn. Reson. Med., vol. 77, no. 4, pp. 1473–1484, 2017.
[5] L. Feng et al., “5D whole-heart sparse MRI,” Magn. Reson. Med., vol. 79, no. 2, pp. 826–838, 2018.
[6] A. Bustin, G. Lima da Cruz, O. Jaubert, K. Lopez, R. M. Botnar, and C. Prieto, “High-dimensionality undersampled patch-based reconstruction (HD-PROST) for accelerated multi-contrast MRI,” Magn. Reson. Med., vol. 81, no. 6, pp. 3705–3719, 2019.
[7] M. R. Prince, H. L. Zhang, S. G. Chabra, P. Jacobs, and Y. Wang, “A pilot investigation of new superparamagnetic iron oxide (ferumoxytol) as a contrast agent for cardiovascular MRI.,” J. Xray. Sci. Technol., vol. 11, no. 4, pp. 231–40, 2003.
[8] Z. Wang, A. C. Bovik, H. R. Sheikh, and E. P. Simoncelli, “Image quality assessment: From error visibility to structural similarity,” IEEE Trans. Image Process., vol. 13, no. 4, pp. 600–612, 2004.
[9] R. Ahmad, Y. Ding, and O. P. Simonetti, “Edge Sharpness Assessment by Parametric Modeling: Application to Magnetic Resonance Imaging,” Concepts Magn. Reson. Part A. Bridg. Educ. Res., vol. 44, no. 3, pp. 138–149, May 2015.
[10] A. Etienne, R. M. Botnar, A. M. C. Van Muiswinkel, P. Boesiger, W. J. Manning, and M. Stuber, “‘Soap-Bubble’ visualization and quantitative analysis of 3D coronary magnetic resonance angiograms,” Magn. Reson. Med., vol. 48, no. 4, pp. 658–666, 2002.
[11] D. Piccini et al., “Deep Learning to Automate Reference-Free Image Quality Assessment of Whole-Heart MR Images,” Radiol. Artif. Intell., vol. 2, no. 3, p. e190123, 2020.
Figures