0125
Automated motion correction of multi-slice fetal brain MRI using a deep recursive framework1Key Laboratory for Biomedical Engineering of Ministry of Education, Department of Biomedical Engineering, College of Biomedical Engineering & Instrument Science, Zhejiang University, Hangzhou, China, 22. Department of Biomedical Engineering, Johns Hopkins University School of Medicine, Baltimore, MD, United States, 3School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai, China, 4Department of Radiology, Shandong Medical Imaging Research Institute, Cheeloo College of Medicine, Shandong University, Jinan, China
Synopsis
Prenatal MRI of fetal brain is vulnerable to unpredictable fetal motion and maternal movement. The conventional registration-based motion correction methods sometimes fail in excessive motion. In this work, we proposed a learning-based scheme to estimate fetal brain motion using a deep recursive framework, which replicated the iterative slice-to-volume registration and 3D volumetric reconstruction process. The network outperformed the previous learning-based methods and with good computational efficiency compared to traditional method. It also achieved high super-resolution reconstruction accuracy on simulated motion-corrupted slices, and therefore, is promising for fetal brain MRI analysis.
Introduction
Fetal MR neuroimaging is usually undermined by severe, uncontrollable, and arbitrary fetal motion, imposing enormous challenges for fetal brain 3D volumetric reconstruction and disease detection1-2. Conventional motion correction and reconstruction of multi-slice T1/T2-weighted fetal brain MRI involves slice-to-volume registration (SVR) and super-resolution reconstruction (SRR) of multi-oriented 2D multi-slice images to solve an inverse problem3-5. However, the method is susceptible to rapid large-scale motion and is excessively time-consuming, which is impractical in clinical settings. In this work, a deep recursive framework (DeepRF) was proposed by mimicking iterations between rigid SVR and 3D reconstruction using deep learning. We compared the method to several state-of-the-art learning-based fetal motion estimation algorithms using simulated motion-corrupted data.Methods
Data acquisition: In-utero MRI scans of 95 fetal brain MRI (gestational age (GA): 23.3-40.0 weeks) with at least three orthogonal orientations were collected under IRB approval. The data were acquired on a 3T Siemens Skyra scanner (Siemens Healthineers, Erlangen, Germany) with an abdominal coil, using T2-weighted half-Fourier single-shot turbo spin-echo (HASTE) sequence with the following protocol: TR/TE = 800/97 ms, in-plane resolution = 1.09 × 1.09 mm, FOV = 256 × 200 mm, 35-55 slices with slice thickness = 2 mm, GRAPPA factor = 2.Data preprocessing: Bias field was corrected using the N4 algorithm6. Then, in-plane motion correction and SRR with an isotropic spacing (0.8mm) were automatically performed via NiftyMIC toolkit7. Fetal brain was extracted by manual masking, followed by 3D non-local means denoising8 and intensity normalization. Next, reconstructed fetal brain volumes were rigidly registered to the corresponding templates9, center-aligned and resized into 144*144*120 by padding zeros in the surrounding regions. Note that the super-resolution reconstructed volume from 2mm slices was used as ground truth.
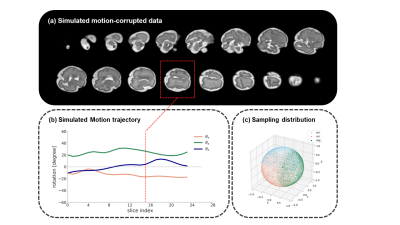
Motion simulation: Fetal brain movement was simulated by 3D rigid transformation. Here, we primarily considered rotational motion, and the rotation angle was sampled from a continuous motion curve generated from a random Markov process and spline interpolation, with the mean between [-45, 45] degrees (Figure 1b). The average velocity of motion was simultaneously regulated by control points. The motion was separated applied to axial, coronal and sagittal orientations, and therefore, the simulated motion covered all possible poses (Figure 1c). In this work, we temporarily simulated non-interleaved acquisition that covered the entire fetal brain with 4 mm slice-thickness.
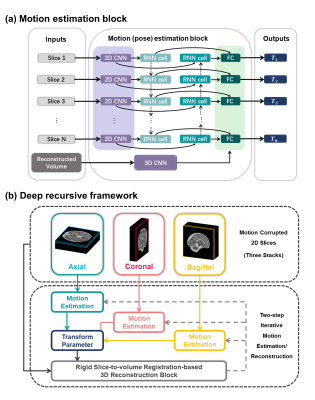
Network architecture: The network is illustrated in Figure.2. For motion estimation, the slices $$$I_s$$$ were fed into a 2D CNN for feature extraction and then went through a bidirectional recurrent network followed by fully-connected layers. The predicted parameters were subsequently utilized for motion correction and 3D reconstruction. We employed a scattered data approximation10 with decreasing Gaussian spatial blurring as the reconstruction transformer that facilitated the network computation. A roughly reconstructed volume $$$V$$$ from motion-corrected axial/coronal/sagittal data was taken back into a 3D CNN and then concatenated to the fully-connected layers. The network weights were shared across iterations and optimized using an end-to-end scheme. The loss $$$L$$$ includes the MSE of rigid transform parameters and the similarity between the estimated images and ground truth. $$L = {L_{sim}}({\hat \phi _s},{\phi _s}) + \lambda {L_{sim}}\left( {{\rm{\Omega }}\left( {{I_s},{{\hat \phi }_s}} \right),V} \right)$$ where $$$L_{sim}$$$ denotes the similarity metric, i.e., MSE, $$${\phi _s}$$$ and $$${\hat \phi _s}$$$ are the rotational angle and its estimation, $$${\rm{\Omega }}$$$ denotes 3D reconstruction, and the two loss terms are adjusted by a weighting $$$\lambda$$$.
Altogether, 82 (GA: 23.3-40.0w), 5 (GA: 26.1-32.0w), and 8 (GA: 24.6-35.4w) fetuses were used for training, validation and testing. The performance of DeepRF was compared to other state-of-the-art learning-based fetal brain motion estimation methods, including Deep Pose Estimation11, anchor-point-based SVRnet12, Deep Predictive Motion Tracking13.
Results
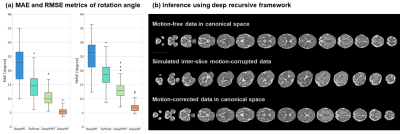
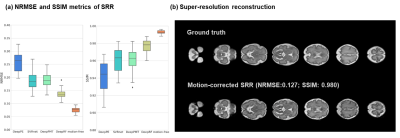
As shown in Figure 3a, the DeepRF achieved the least mean absolute error (MAE) of 5.49º and root MSE of 7.23º, compared to other learning-based methods on the simulated motion-corrupted slices. The average time for motion correction is 15.9 seconds, with nearly 50-fold speed acceleration compared to the conventional two-step iterative SVR-SRR method.We performed SRR on the motion-corrected data from different networks, and DeepRF achieved the best reconstruction accuracy with the least normalized root mean square error (NRMSE) of 0.136 and the highest structural similarity index (SSIM) of 0.977 with the ground truth (2mm slice-thickness acquisition) (Figure 4), which was close to the SRR of motion-free data with NRMSE of 0.076 and SSIM of 0.993 (4mm slice-thickness acquisition).
Discussion and conclusion
The proposed DeepRF outperformed the other state-of-the-art learning-based methods for fetal brain motion estimation and correction, and also increased the 3D reconstruction efficiency with close precision compared to the traditional SRR scheme. The superior performance of the DeepRF may be associated with the network architecture that replicates the iterative SVR and reconstruction process and incorporation of the image similarity and transform matrices, as well as the use of bidirectional recurrent network that takes into account the series of motion between successive slices. The high accuracy and computational efficiency of DeepRF makes it promising for fetal brain MRI analysis, although generalizability of the current network needs to be further tested in more complicated situations.Acknowledgements
This work is supported by the Ministry of Science and Technology of the People’s Republic of China (2018YFE0114600), National Natural Science Foundation of China (61801424 and 81971606).References
1. Glenn O A, Barkovich A J. Magnetic resonance imaging of the fetal brain and spine: an increasingly important tool in prenatal diagnosis, part 1. American Journal of Neuroradiology, 2006; 27(8): 1604-1611. 2. Malamateniou C, et al. Motion-compensation techniques in neonatal and fetal MR imaging. American Journal of Neuroradiology, 2013; 34(6): 1124-1136.
3. Gholipour A, Estroff J A, Warfield S K. Robust super-resolution volume reconstruction from slice acquisitions: application to fetal brain MRI. IEEE transactions on medical imaging, 2010; 29(10): 1739-1758.
4. Kuklisova-Murgasova M, et al. Reconstruction of fetal brain MRI with intensity matching and complete outlier removal. Medical image analysis, 2012; 16(8): 1550-1564.
5. Marami B, et al. Temporal slice registration and robust diffusion-tensor reconstruction for improved fetal brain structural connectivity analysis. NeuroImage, 2017; 156: 475-488.
6. Tustison N J, et al. N4ITK: improved N3 bias correction. IEEE transactions on medical imaging, 2010; 29(6):1310-1320.
7. Ebner M, et al. An automated framework for localization, segmentation and super-resolution reconstruction of fetal brain MRI. NeuroImage, 2020; 206: 116324.
8. Coupé P, et al. An optimized blockwise nonlocal means denoising filter for 3-D magnetic resonance images. IEEE transactions on medical imaging, 2008; 27(4):425-441.
9. Serag A, et al. Construction of a consistent high-definition spatio-temporal atlas of the developing brain using adaptive kernel regression. NeuroImage, 2012; 59 (3):2255-65.
10. Rousseau F,et al. Registration-based approach for reconstruction of high-resolution in utero fetal MR brain images. Academic radiology, 2006; 13(9):1072-1081.
11. Salehi S S M, Khan S, Erdogmus D, et al. Real-time deep pose estimation with geodesic loss for image-to-template rigid registration. IEEE transactions on medical imaging, 2018; 38(2): 470-481.
12. Hou B, et al. 3-D reconstruction in canonical co-ordinate space from arbitrarily oriented 2-D images. IEEE transactions on medical imaging, 2018; 37(8):1737-1750.
13. Singh A, Salehi S S M, Gholipour A. Deep Predictive Motion Tracking in Magnetic Resonance Imaging: Application to Fetal Imaging. IEEE Transactions on Medical Imaging 2020.
Figures