0100
Whole-Brain 3D Quantitative BOLD Mapping With Preliminary Estimation of R2' and Venous Blood Volume1Radiology, University of Pennsylvania, Philadelphia, PA, United States
Synopsis
Quantitative BOLD (qBOLD) seeks to quantify voxel-wise deoxygenated blood volume (DBV) and venous blood oxygen saturation (Yv) based on R2′-sensitive signal acquisitions. A complication in qBOLD is the separation of signal contributions from R2, and R2′ from heme and non-heme iron sources, particularly in the deep brain structures. Here, we develop a new 3D qBOLD mapping method by tackling the confounding factors based on preliminary estimates of R2, R2’, and voxel susceptibility, along with cerebral venous blood volume. Results suggest feasibility of the proposed, prior-based qBOLD method for 3D mapping of DBV and Yv across the entire brain.
Introduction
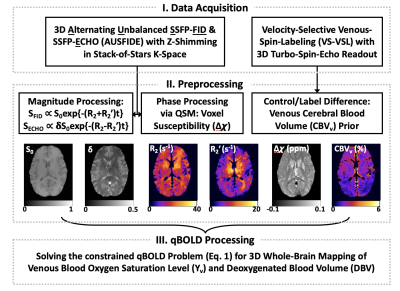
Quantitative BOLD (qBOLD)1,2 seeks to quantify voxel-wise deoxygenated blood volume (DBV) and venous blood oxygen saturation (Yv) based on R2'-sensitive signal acquisitions. However, in the original qBOLD method, it is challenging to separate the two parameters from the estimated R2' because of their mutual coupling in the model3. To address this issue, we had previously proposed to combine extravascular R2' and intravascular R2 mapping approaches for prior information based qBOLD processing4. While the method substantially improved stability of the parameter quantification, it only allows 2D single-slice quantification, thus limiting its utility in clinical practice. A further complication in the qBOLD method is the separation of signal contributions from R2, and R2' from heme and non-heme iron sources, particularly in the deep brain structures (e.g., putamen, globus pallidus). Thus, the objective of this work was to develop a new 3D MRI oximetry technique that enables robust qBOLD parameter mapping across the entire brain. To this end, we implemented a 3D qBOLD processing pipeline, which takes into account the confounding factors by making use of preliminary estimates of R2, R2', and voxel susceptibility (χ) obtained using our recently reported technique, referred to as alternating unbalanced SSFP-FID and SSFP-ECHO (AUSFIDE)5, and additionally measured venous cerebral blood volume (CBVv) via a velocity-selective venous-spin-labeling (VS-VSL) method6.Methods
Pulse sequence & problem definition: The AUSFIDE pulse sequence5 permits rapid 3D encoding of both R2 and R2' by collecting multiple SSFP-FID and SSFP-ECHO signals alternately over sequence repetitions. Furthermore, the method’s inherent sensitivity to large-scale induced magnetic field offsets (ΔB0) was tackled by means of 3D z-shimming and voxel-spread-function (VSF)-based correction7,8. Figure 1 shows the proposed qBOLD processing pipeline, in which the AUSFIDE-derived parameters, R2, R2', and χ, are employed to serve as preliminary estimates in solving the following problem:$$\underset{\Theta}{\arg\min}\sum_{t}\lvert y(t)-\Xi(\Theta,t)\rvert^2+w\lvert \chi-\Psi(\Theta) \rvert^2+p\lvert R_2'-\Gamma(\Theta) \rvert^2 (1) $$
Here, y(t) is the acquired multi-echo AUSFIDE signal at the sampling times t, Ξ is the corresponding model accounting for R2, R2', and ΔB0-induced signal modulations, $$$\Theta=\{Y_v,DBV,R_{2,nb}',\chi_{nb}\}$$$ is the unknown parameter set, ψ is the four-pool decomposition model for χ9,10, Γ disentangles R2' contributions from the non-blood and deoxygenated blood, and w and p are the regularization parameters that enforce prior knowledge in the qBOLD parameter estimation. Exact forms of Ξ, ψ, and Γ along with descriptions for sub-parameters therein are provided in Figure 2. In solving Eq. (1), VS-VSL-derived 3D CBVv maps were employed to initialize and constrain the solution of DBV.
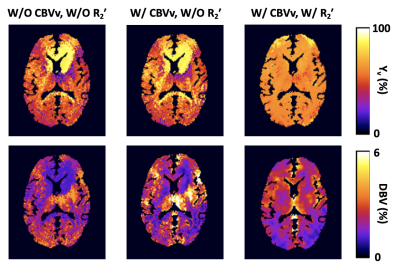
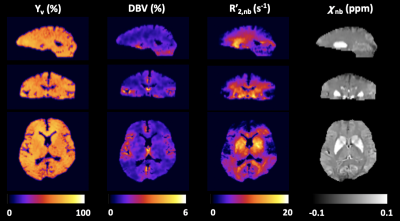
Experiments & data processing: Experiments were performed at 3T (Siemens Prisma) in six healthy subjects. Here, the AUSFIDE data collected in the study of Ref. 5 were used, while the VS-VSL data were newly acquired using the imaging parameters specified in Ref. 6. High-resolution T1-weighted images were additionally acquired for brain segmentation. To evaluate the effectiveness of providing prior information of R2' and CBVv in solving the qBOLD problem, Eq. (1) was solved for an axial slice of a subject in the following three different ways: 1) p=0 and no CBVv prior, 2) p=0 and CBVv prior, and 3) p>0 and CBVv prior, leading to Yv and DBV maps, respectively. For the cases of 1) and 2), DBV was initialized with 2%, while for 3) DBV in each pixel was initialized to CBVv with the solution bound: [0.5CBVv, 2CBVv]. χ was measured using the MEDI toolbox11, and Yv was initialized based on the χ values in the superior sagittal sinus. Hematocrit was measured individually for each subject, and was scaled with a factor of 0.85 for microvascular compartments12. Derived 3D maps of Yv, DBV, R'2,nb, and χnb in a subject were presented in sagittal, coronal, and axial orientations. Group averages of Yv and DBV across the six study subjects were calculated in gray and white matter regions.
Results
Figure 3 shows pairs of Yv and DBV maps in the axial plane, derived in three different ways of using prior estimates of R2' and CBVv. Only the case employing both priors yields physiologically plausible Yv (spatially nearly uniform) and DBV (gray and white matter contrast with the values in the former larger than those in the latter) maps. Figure 4 displays 3D maps of Yv, DBV, R'2,nb, and χnb in the three orthogonal planes. Group-averages of Yv and DBV are Yv=64.1±1.7% and 63.7±1.7%, and DBV=2.6±0.6% and 1.7±0.5% for gray and white matter regions, respectively.Discussion and Conclusions
We introduced a new MRI oximetry method that enables 3D qBOLD parameter mapping across the entire brain. The results show the expected contrast in all four parameter maps obtained, i.e., a clear depiction of gray/white matter DBV difference, relatively homogeneous Yv distribution. The data further highlight elevated R'2,nb and χnb in the iron-rich basal ganglia, relative to cortical regions, thus suggesting that the present method making use of preliminary estimates of R2' and CBVv achieves robust separation of several sources that contribute to voxel signal modulations in the brain. The proposed technique may find a range of clinical applications where oxygen extraction is elevated (such as in regional brain ischemia). Finally, in combination with cerebral blood flow imaging, the method should allow 3D voxelwise mapping of the cerebral metabolic rate of oxygen consumption across the entire brain.Acknowledgements
This work was supported by the National Institutes of Health grant R21EB022687 and the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR001878.References
1. An HY, Lin WL. Quantitative measurements of cerebral blood oxygen saturation using magnetic resonance imaging. J Cereb Blood Flow Metab 2000; 20:1225-1236.
2. He X, Yablonskiy DA. Quantitative BOLD: mapping of human cerebral deoxygenated blood volume and oxygen extraction fraction: default state. Magn Reson Med. 2007;57:115-126.
3. Sedlacik J, Reichenbach JR. Validation of quantitative estimation of tissue oxygen extraction fraction and deoxygenated blood volume fraction in phantom and in vivo experiments by using MRI. Magn Reson Med. 2010;63:910-921.
4. Lee H, Englund EK, Wehrli FW. Interleaved quantitative BOLD: Combining extravascular R2’- and intravascular R2-measurements for estimation of deoxygenated blood volume and hemoglobin oxygen saturation. NeuroImage. 2018;174:420-431.
5. Lee H, Wehrli FW. Alternating unbalanced SSFP for 3D R2’ mapping of the human brain. Magn Reson Med. 2020. DOI: https://doi.org/10/1002/mrm.28637.
6. Lee H, Wehrli FW. Venous cerebral blood volume mapping in the whole brain using venous-spin-labeled 3D turbo spin echo. Magn Reson Med. 2020;84:1991-2003.
7. Yablonskiy DA, Sukstanskii AL, Luo J, Wang X. Voxel spread function method for correction of magnetic field inhomogeneity effects in quantitative gradient-echo-based MRI. Magnetic Resonance in Medicine 2013;70:1283-1292.
8. Han D, Nam Y, Gho SM, Kim DH. Volumetric R2* mapping using z-shim multi-echo gradient echo imaging. Magnetic Resonance in Medicine 2015;73:1164-1170.
9. Zhang J, Liu T, Gupta A, Spincemaille P, Nguyen TD, Wang Y. Quantitative mapping of cerebral metabolic rate of oxygen (CMRO2) using quantitative susceptibility mapping (QSM). Magn Reson Med. 2015;74(4):945-952.
10. Cho J, Kee Y, Spincemaille P, Nguyen TD, Zhang J, Gupta A, et al. Cerebral metabolic rate of oxygen (CMRO2) mapping by combining quantitative susceptibility mapping (QSM) and quantitative blood oxygenation level‐dependent imaging (qBOLD). Magn Reson Med. 2018;80(4):1595-604.
11. Liu J, Liu T, de Rochefort L, Ledoux J, Khalidov I, Chen W, Tsiouris AJ, Wisnieff C, Spincemaille P, Prince MR. Morphology enabled dipole inversion for quantitative susceptibility mapping using structural consistency between the magnitude image and the susceptibility map. Neuroimage 2012;59(3):2560-2568.
12. Eichling JO, Raichle ME, Grubb RL, Larson KB, Ter-Pogossian MM. In vivo determination of cerebral blood volume with radioactive oxygen-15 in the monkey. Circ Res. 1975;37:707-714.
Figures