0097
Construction of Spatiotemporal Cortical Surface Atlases for Fetal Brains1Department of Radiology and BRIC, UNC-Chapel Hill, Chapel Hill, NC, United States, 2Department of Radiology, Obstetrics and Gynecology Hospital, Fu Dan University, Shanghai, China
Synopsis
We constructed a set of temporally-densely sampled cortical surface atlases for the fetal brain from 22 to 36 gestational weeks. This 4D fetal cortical surface atlas, which will be released to the public soon, together with the UNC 4D Infant Cortical Surface Atlas provide the longest temporally-consistent atlas chain from the prenatal 22 gestational weeks to the postnatal 7 years of age.
Introduction
Spatiotemporal (4D) fetal cortical surface atlases are critical for visualization, spatial normalization and analysis of the dynamically expanding fetal brain cortex, but remain still scarce. To fill this gap, in this paper, we present a densely-sampled 4D cortical surface atlas, including 9 time points from 22 to 36 gestational weeks, based on fetal brain MR images from 98 healthy subjects. To construct the atlas at each time point, we first conduct the unbiased co-registration of the cortical surfaces within the same age group to obtain the age-specific group-mean surface. Then, we align these group-mean surfaces to the common space, i.e., UNC 4D Infant Cortical Surface Atlas at the neonatal time-point [1], by subsequently aligning the group-mean surfaces between neighboring time points in an age-descent order, thus ensuring both temporal consistency and unprecedented continuity from the fetal atlas to the infant atlas. These fetal cortical surface atlases are therefore served as a complementary to our released UNC 4D Infant Cortical Surface Atlas, for unprecedentedly providing the longest temporally-consistent atlas chain from the prenatal 22 gestational weeks to the postnatal 7 years of age.Materials and Methods
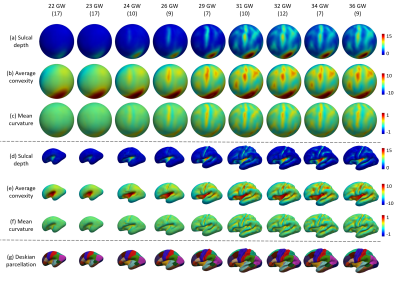
In this study, brain MR images collected from 98 healthy fetuses during 22 to 36 gestational weeks were used for the cortical surface atlases construction at 9 time points, i.e., 22, 23, 24, 26, 29, 31, 32, 34 and 36 gestational weeks. The subject number included in each age group was presented in Fig. 1.T2-weighted MRI stacks of fetuses were acquired by a 1.5 T Siemens Avanto scanner with the resolution of 0.54x0.54x4.4 mm3. Brain localization, extraction [2], and super-resolution volume reconstruction from 2D stacks were performed to generate the 3D brain volume with an isotropic resolution of 0.8 mm [3]. Brain tissues were segmented into the white matter, gray matter and cerebrospinal fluid [4] and then be manually corrected. Then, the topologically correct and geometrically accurate cortical surfaces of each hemisphere were reconstructed using an in-house fetal/infant cortical surface analysis pipeline [5]. With the reconstructed cortical surfaces, the vertex-wise cortical attributes for measuring the cortical folding, including the sulcal depth, average convexity, and mean curvature, were computed, which were used for aligning the cortical surfaces across different subjects.
To facilitate the cortical surface alignment across different subjects, the inner cortical surfaces were inflated and mapped onto a sphere with the minimal geometric distortion [6]. Then, the spherical cortical surfaces within the same age group were aligned together using an unbiased group-wise registration [7] and the co-registered spherical surfaces were resampled with a uniform spherical mesh tessellation, resulting in a vertex-wise correspondence across different subjects in the same age group. Then, the average cortical surface and folding attributes were obtained by averaging the corresponding vertices’ positions and their attributes, leading to an un-biased and age-specific group mean surface at each time point.
To establish the spatial correspondences between the fetal atlases and the released UNC 4D Infant Cortical Surface Atlas, the group-mean surface at the 36 gestational weeks was first aligned onto the neonatal atlas in UNC 4D Infant Cortical Surface Atlas [1]. Then, for every two neighboring time points, the group-mean surface at the younger age was aligned to the group-mean surface at the older age in an age-descent order, until all group-mean surfaces were aligned. This strategy can better preserve the temporal consistency of the atlases at different ages. Finally, these aligned group-mean surfaces were resampled as the atlases at the specific time-points. For equipping our fetal atlases with the meaningful parcellations, we thereby propagated the Desikan parcellation [8] from the infant cortical surface atlas [1] to our fetal atlas, since there were vertex-to-vertex correspondences between the fetal and infant cortical surface atlases.
Results
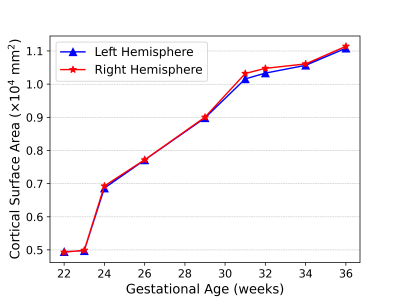
Fig. 1 shows the constructed spatiotemporal fetal cortical surface atlases with the color-coded sulcal depth, average convexity and mean curvature at each time point on both the spherical surface and the average inner cortical surface. From Fig. 1, we can see that there is rapid cortex development from 22 to 36 gestational weeks. Specifically, the cortex expands dramatically and the cortical folding degree increases remarkably. At the 36 gestation weeks, we have already seen the major cortical folds on the cortex. To better show the cortex expansion, Fig. 2 shows the average cortical surface area (without subcortical) at different time points. From the figure, the cortical surface area expands 120% from the 22 to 36 gestational weeks.Conclusions
We constructed a set of temporally-densely sampled cortical surface atlases for the fetal brain from 22 to 36 gestational weeks. This 4D fetal cortical surface atlas, which will be released to the public soon, together with the UNC 4D Infant Cortical Surface Atlas provide the longest temporally-consistent atlas chain from the prenatal 22 gestational weeks to the postnatal 7 years of age. These 4D fetal and infant surface atlases will be unique and valuable resources for consistently studying the most dynamic stages of prenatal and postnatal brain development.Acknowledgements
This work was partially supported by NIH grants (MH116225, MH117943).References
[1] Z. Wu, L. Wang, W. Lin, J. H. Gilmore, G. Li, and D. Shen, “Construction of 4D infant cortical surface atlases with sharp folding patterns via spherical patch-based group-wise sparse representation,” Hum. Brain Mapp., vol. 40, no. 13, pp. 3860–3880, 2019.
[2] L. Liao et al., “Joint Image Quality Assessment and Brain Extraction of Fetal MRI Using Deep Learning,” in International Conference on Medical Image Computing and Computer-Assisted Intervention, 2020, pp. 415–424.
[3] M. Ebner et al., “An automated framework for localization, segmentation and super-resolution reconstruction of fetal brain MRI,” Neuroimage, vol. 206, p. 16324, 2020.
[4] Y. Pei et al., “Anatomy-Guided Convolutional Neural Network for Motion Correction in Fetal Brain MRI,” in International Workshop on Machine Learning in Medical Imaging, 2020, pp. 384–393.
[5] G. Li, L. Wang, F. Shi, W. Lin, and D. Shen, “Constructing 4D infant cortical surface atlases based on dynamic developmental trajectories of the cortex,” in International Conference on Medical Image Computing and Computer-Assisted Intervention, 2014, pp. 89–96.
[6] B. Fischl, M. I. Sereno, and A. M. Dale, “Cortical surface-based analysis: II. Inflation, flattening, and a surface-based coordinate system,” Neuroimage, vol. 9, no. 2, pp. 195–207, 1999.
[7] B. T. T. Yeo, M. R. Sabuncu, T. Vercauteren, N. Ayache, B. Fischl, and P. Golland, “Spherical demons: Fast diffeomorphic landmark-free surface registration,” IEEE Trans. Med. Imaging, vol. 29, no. 3, pp. 650–668, 2010.
[8] R. S. Desikan et al., “An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest,” Neuroimage, vol. 31, no. 3, pp. 968–980, 2006.
Figures