0060
Predicting Gadolinium Contrast Enhancement for Structural Lesion Analysis using DeepContrast1Department of Biomedical Engineering, Columbia University, New York, NY, United States, 2VantAI, New York, NY, United States, 3Department of Biological Sciences and the Taub Institute, Columbia University, New York, NY, United States, 4Department of Electrical Engineering and the Taub Institute, Columbia University, New York, NY, United States, 5Department of Neurology, the Taub Institute, the Sergievsky Center, Radiology and Psychiatry, Columbia University, New York, NY, United States, 6Department of Psychiatry, Mortimer B. Zuckerman Mind Brain Behavior Institute, Columbia University, New York, NY, United States
Synopsis
Gadolinium-based contrast agents (GBCAs) have facilitated an improved analysis and understanding of structural lesions, however, present safety risks due to the tissue retention of GBCAs. Here we optimize and apply the deep learning model, DeepContrast, to predict gadolinium uptake in brain and breast structural lesions for structural lesion enhancement. The optimized DeepContrast models predict gadolinium uptake that is comparable to ground-truth scans consisting of the uptake from the GBCAs, using a single T1-weighted pre-contrast scan.
Introduction
For the detection and subsequent enhancement of structural lesions, gadolinium-based contrast agents (GBCAs) can be administered with MRI.1 For brain lesions such as brain tumors, where there is frequently a breakdown of the blood-brain barrier,2 these contrast agents can assist in improving lesion characterization.3 Similar benefits have been seen with breast tumors, as GBCAs allow for the improved characterization and vascular visualization of these lesions.4 While the diagnostic benefits of GBCAs are significant, the safety and long-term health risks of GBCAs are becoming a growing concern.5 Specific concerns are around the retention of GBCAs, especially within the brain, which may lead to retention-created toxicity.6.7 Preserving the structural lesion diagnostic utility of GBCAs while removing the safety concern that they present is a task that can be accomplished through deep learning. Studies have shown that gadolinium can be reduced while preserving contrast information,8 while others have used multiparametric MRI for contrast prediction.9 In this study, we use our previously developed deep learning model DeepContrast,10 to predict the enhancement of brain and breast structural lesions using a single T1-weighted (T1W) pre-contrast scan.Methods
Brain Tumor Data PreprocessingThe Brain Tumor Segmentation (BraTS) dataset was used for brain lesions.11 Data for 128 subjects were used, based on successful tissue segmentation and data cleaning. Each subject included a T1W, T1W-CE, and tumor region segmentations. Scans for all subjects were registered to a template space using rigid registration and N4 bias correction was applied using ANTS.12 T1W scan normalization was completed using the largest intensity of the non-tumor region. T1W-CE scans were normalized through scaling correction, using a scaling ratio calculated with average intensities of the white matter regions within the TW1 and TW1-CE scans. This scaling correction was done to ensure the T1 pairs are in the same scaling system, a property needed to determine gadolinium uptake. This can be accomplished using a region of the brain which shows minimal change after contrast enhancement, a behavior demonstrated by white matter.13 The pairs were randomly distributed into train-test-validation splits, consisting of 101, 15, and 12 pairs of scans respectively.
Brain Tumor Model Training
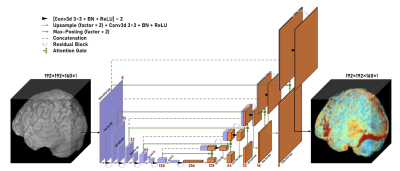
As the scans were originally acquired through 3D acquisition with isotropic voxel spacing, a 3D Residual Attention U-Net model was used during training (Figure 1). The package TorchIO14 was used to introduce image augmentation during training, to assist with model generalization. The model was trained using the 3D brain volumes and the ground truth gadolinium uptake was defined as the difference between the T1W contrast-enhanced (T1W-CE) and pre-contrast scans.
Breast Tumor Data Preprocessing
The Breast-MRI-NACT-Pilot dataset15 was used for breast lesions which included data from 68 subjects. The imaging protocol included a 3D localizer and a unilateral sagittal DCE acquisition.16 For each DCE acquisition, a pair of scans were used including the non-contrast T1W scan and the scan acquired at the initial time point of the DCE protocol, which was used as the T1W-CE scan. This resulted in 161 pairs of scans, which were randomly distributed into train-test-validation splits consisting of 129, 16, and 16 pairs respectively. Tumor masks were processed to ensure proper spatial matching with the non-contrast and contrast-enhanced scans. K-means clustering was used to generate whole breast masks.
Breast Tumor Model Training
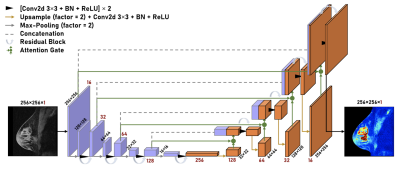
A 2D Residual Attention U-Net model was used during training (Figure 2). The model was trained using 2D sagittal breast scan slices where the ground truth gadolinium uptake was defined as the difference between the T1W-CE scan and the T1W scan
Results
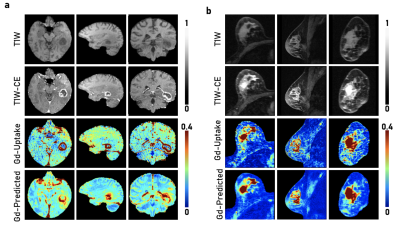
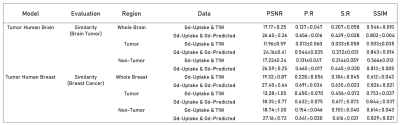
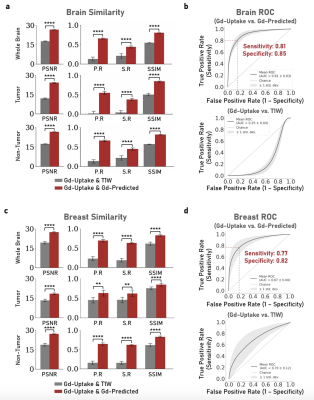
For both the brain and breast lesions, the optimized models were evaluated on their respective hold-out sets by evaluating metrics on the tumor regions specified by the tumor masks, as well as through voxel-wise analysis across the whole organ region. Figure 3 shows sample test scans, demonstrating the qualitative comparability and concordance between the ground truth GBCA uptake of the structural lesions and the contrast level estimated by model predictions. Visually, breast lesion predictions show superior enhancements compared to brain lesions around smaller vessels. Figure 4 summarizes the PSNR, Pearson R Correlation, Spearman R Correlation, and SSIM, calculated between model predictions and ground truth data. Figure 5 summarizes the AUC, Sensitivity, and Specificity and shows the receiver operator characteristic (ROC) curves, generated by evaluating 1000 different binarizing thresholds. These metrics highlight the predictive potential of DeepContrast for both brain and breast structural lesion enhancement, from a single T1W pre-contrast scan.Conclusions and Discussion
Results demonstrated that DeepContrast can predict structural lesion contrast enhancements which are qualitatively and quantitatively comparable to the ground truth GBCA uptake. This is significant given the high variability of structural lesions, such as glioblastomas, which demonstrate significant variation in shape and form.17 The significance of these results is further demonstrated given that the predicted contrast is from a single T1W pre-contrast scan. While our results show the potential of replacing GBCAs or significantly reducing their dosage when imaging structural lesions, they can be further improved given more data that better encompasses the high variability of structural lesions to improve prediction of finer details, specifically within the brain.Acknowledgements
This work was supported, in part, by the National Institute of Health and was also performed at Zuckerman Mind Brain Behavior Institute MRI Platform, a shared resource and Columbia MR Research Center site.References
1. Lohrke, J. et al. 25 years of contrast-enhanced mri: developments, current challenges and future perspectives. Advances in therapy 33, 1–28 (2016).
2. Borges, A., López-Larrubia, P., Marques, J. & Cerdan, S. Mr imaging features of high-grade gliomas in murine models: how they compare with human disease, reflect tumor biology, and play a role in preclinical trials. American journal of neuroradiology 33, 24–36 (2012).
3. Shen, Q. & Duong, T. Q. Magnetic resonance imaging of cerebral blood flow in animal stroke models. Brain circulation 2, 20 (2016).
4. Lenkinski, R. E., Wang, X., Elian, M., & Goldberg, S.N. Interaction of gadolinium-based MR contrast agents with choline: Implications for MR spectroscopy (MRS) of the breast. Magnetic Resonance in Medicine 61, 1286–1292 (2009)
5. Quattrocchi, C. C. & van der Molen, A. J. Gadolinium retention in the body and brain: is it time for an international joint research effort? (2017)
6. Tedeschi, E., Caranci, F., Giordano, F., Angelini, V., Cocozza, S., & Brunetti, A. Gadolinium retention in the body: what we know and what we can do. Radiologia Medica, 122, 589–600 (2017)
7. Ramalho, J., Ramalho, M., Jay, M., Burke, L.M., & Semelka, R. C. Gadolinium toxicity and treatment (2016)
8. E. Gong, J. M. Pauly, M. Wintermark, & G. Zaharchuk. Deep learning enables reduced gadolinium dose for contrast enhanced brain MRI. Journal of Magnetic Resonance Imaging 48, 330-340 (2018).
9. J. Kleesiek et al. Can Virtual Contrast Enhancement in Brain MRI Replace Gadolinium?: A Feasibility Study. Investigative Radiology, 54, 653-660 (2019).
10. H. Sun et al. Substituting Gadolinium in Brain MRI Using DeepContrast, 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 2020, pp. 908-912 (2020)
11. Menze, B. H. et al. The multimodal brain tumor image segmentation benchmark (brats). IEEE transactions on medical imaging 34, 1993–2024 (2014).
12. Avants, B. B., Tustison, N. & Song, G. Advanced normalization tools (ants). Insight j 2, 1–35 (2009).
13. Feng, X. et al. Temporal lobe epilepsy lateralization using retrospective cerebral blood volume mri. NeuroImage: Clinical 19, 911–917 (2018)
14. Pérez-García, F., Sparks, R. & Ourselin, S. Torchio: a python library for efficient loading, preprocessing, augmentation and patch-based sampling of medical images in deep learning (2020). 2003.04696.
15. Clark, K. et al. The cancer imaging archive (tcia): maintaining and operating a public information repository. Journal of digital imaging 26, 1045–1057 (2013).
16. Newitt, D. & Hylton, N. Single site breast dce-mri data and segmentations from patients undergoing neoadjuvant chemotherapy. The Cancer Imaging Archive 2 (2016)
17. Georgescu, M.M., & Olar, A. Genetic and histologic spatiotemporal evolution of recurrent, multifocal, multicentric and metastatic glioblastoma. Acta Neuropathologica Communications 8, 10 (2020)
Figures