S43
Assessing the reproducibility of semi-automated segmentation methods on post-operative T1 post contrast and FLAIR MRI images of GBM1Imaging, Imperial College Healthcare NHS Trust, London, United Kingdom, 2Imperial Health Charity, London, United Kingdom, 3Department of Surgery & Cancer, Cancer Imaging Centre, Imperial College London, London, United Kingdom, 4Centre for Clinical Brain Sciences, University of Edinburgh, Edinburgh, United Kingdom
Synopsis
Glioblastomas (GBMs) are common adult primary brain tumours with high variable prognosis and a therapeutic response and outcome dominated by individual tumour biology. Magnetic resonance imaging (MRI) allows assessment of disease status following treatment. Accurate segmentation of GBM is the key for development of novel imaging biomarkers. A total of 35 MRI scans were analysed from patients withbiopsy confirmed GBM. Segmentation reproducibility between the two readers using semi automation was assessed and was found to be borderline. Future work is warranted using fully automated techniques to refine this ongoing work.
BACKGROUND AND PURPOSE
Glioblastoma (GBM) account for ~70% of adult primary brain tumours [1]. Despite optimal current treatment, median survival remains only 12-15 months; prognosis is also highly variable; therapeutic response and outcome is dominated by individual tumour biology [2,3,4]. Magnetic Resonance imaging (MRI) is widely used in assessment of disease status following maximal surgical resection and chemoradiotherapy. Accurate segmentation of GBM is the key for development of novel imaging biomarkers, as well as driving more computationally advanced methods for disease detection and prognostication e.g. Radiomics. In clinical application, radiologists will perform manual segmentation, which is subjective, time consuming and difficult to reproduce. Semi- automated segmentation methods can improve efficiency [5] although there is lack of evidence surrounded the reproducibility of these segmentations due to variables in seed points, thresholding values and iteration termination conditions, and variation in subjective manual editing that is performed to correct the automated masks. Moreover, manually edited segmentations are frequently used as ‘ground truth’ in evaluation of the reliability of fully automated intensity and AI based methods.The purpose of our study is to assess the reproducibility of semi-automated segmentation techniques on post-operative T1 contrast and FLAIR sequences in patients with GBM.METHODS
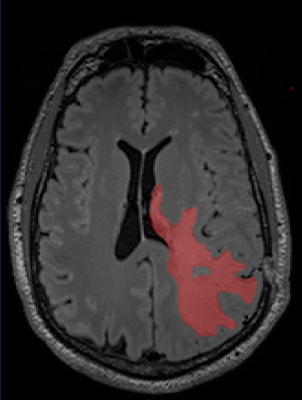
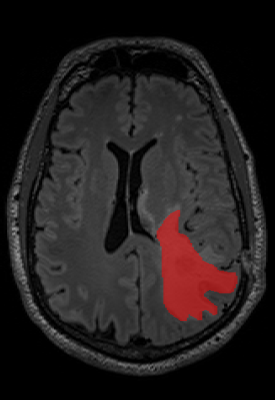
A total of 35 MRI examinations were analysed from patients with biopsy confirmed GBM. All imaging was performed using a Verio (3T) (Siemens, Erlangen, De). The MRI protocol included a volumetric T2- weighted FLAIR sequence (TR 5000ms, TE 329 ms) and a volumetric T1-weighted contrast enhanced (TR 1900ms, TE 2.5ms). Solid enhancing tumour residuum on the T1-weighted post contrast images, and abnormal signal on the T2-weighted FLAIR sequences using commercially available freeware, ITK-snap, using a semiautomated thresholding approach independently by two readers. The semi-automated method involved manual seeding points, iterative automated intensity-based growth of regions and manual correction at the end. The first reader was a MRI specialized radiographer with 12 years’ experience and the second was a radiologist with 9 years of clinical experience. Segmentation reproducibility between the two readers was assessed by calculating the Sorenson-Dice coefficients.RESULTS
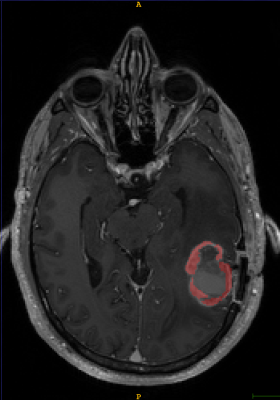
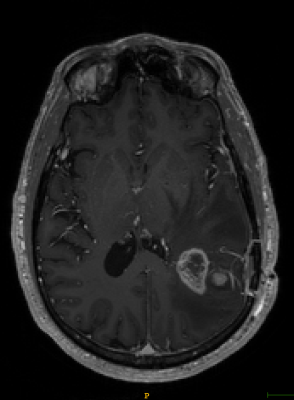
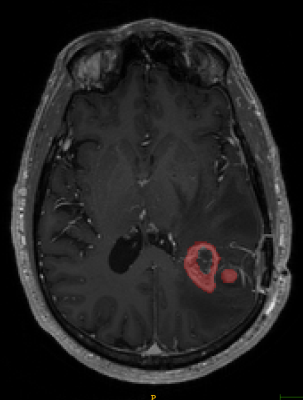
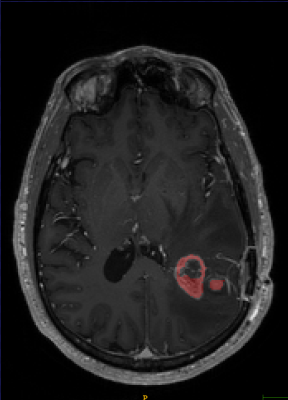
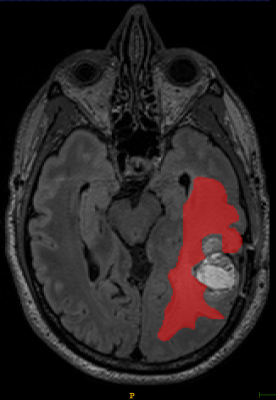
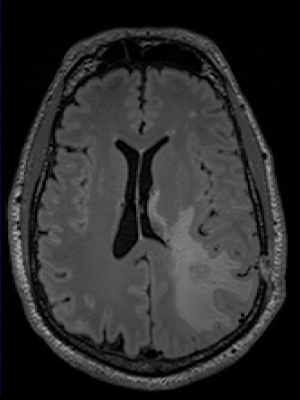
The Sorenson-Dice coefficient for the T1-weighted image segmentation was 0.58±0.19 and for the T2-weighted FLAIR segmentation was 0.67±0.19. Representative figures (1-12) showing axial brain MR images at same slice location with segmentation performed by two MR readers are shown below for both pulse sequences.CONCLUSION
This is the first time the reproducibility of semi-automated segmentation techniques have been tested in post-operative GBM patients. The Dice Coefficient was lower than expected and are clearly not reproducible enough to produce ‘ground truth’ segmentations. The low Dice coefficients would have been confounded by firstly the experience of the two readers in interpreting disease on the two sequences. GBM is notoriously heterogenous on MRI, and although only solid enhancing components were included int the segmentation, it’s is often open to interpretability between readers, where the imaging appearances don’t fall in to either category. Semi-automated region-based techniques used in this study obtain the segmentation result by iteratively adding adjacent to the specified seed points. The disadvantage is that the seed points need to be specified manually which again introduces human bias. At present in our study semi-automated techniques have failed to be reproducible to the level at which the segmentations can be considered ‘ground truth’. This can have important implications for use as reference standards for validation of new automation methods. Future work will include further investigation of causes of variance, and comparisons with fully automated techniques.Acknowledgements
Olga Costa research is supported by Imperial Health Charity, London, United KingdomReferences
[1] Wen PY et al. Malignant gliomas in adults. N Eng J Med. 2008; 359-492.http://www.ncbi.nlm.nih.gov/pubmed/18669428
[2] Lassman AB et al. Incorporating molecular tools into clinical trials and treatment for gliomas? Curr Opin Neurol. 2007, 20:708–711. http://www.ncbi.nlm.nih.gov/pubmed/17992094
[3] Louis DN. Molecular pathology of malignant gliomas. Annu. Rev. Pathol. Mech. Dis. 2006, 1:97–117.
[4] Scheithauer BW et al. The 2007 WHO classification of tumors of the nervous system: controversies in surgical neuropathology. Neuropathology Brain Pathology. 2008, 18:307–316. http://www.brainlife.org/reprint/2008/Scheithauer_BW080602.pdf
[5] (Wu, Zhao, Wu, Lin, & Wang, 2019)Wu, Y., Zhao, Z., Wu, W., Lin, Y., & Wang, M. (2019) Automatic glioma segmentation based on adaptive superpixel. BMC Medical Imaging, 19(1), 1–14. https://doi.org/10.1186/s12880-019-0369-6
Figures
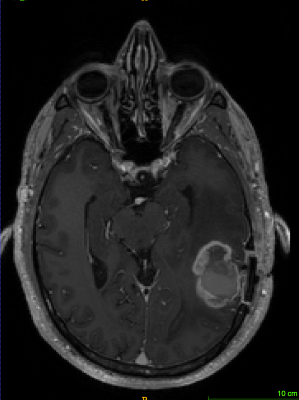
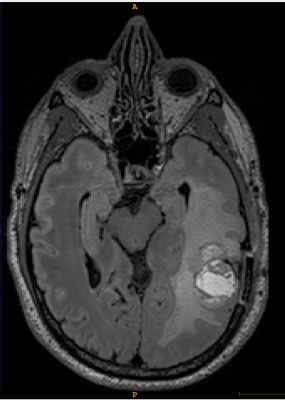
T1-weighted axial contrast enhanced MRI brain image showing GBM lesion before the segmentation (Fig 1)
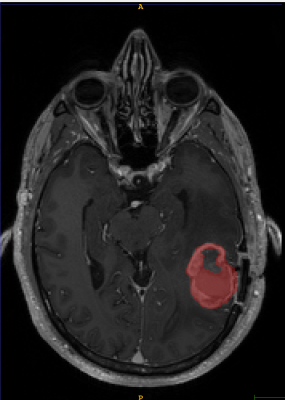
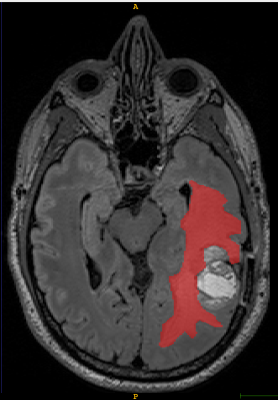
T1-weighted axial contrast enhanced MRI brain image showing GBM lesion after the first reader segmentation (Fig 2)