Utility of Susceptibility Map-Weighted Imaging for Enhanced Visual Identification of Subthalamic Nucleus at 3T: A Feasibility Study
Weiling Lee1, Peik Yen Teh1, Hartono Septian2,3, Nicolas Kon Kam King4, Prakash Kumar M 2, Jongho Lee5, and Ling Ling Chan1,3
1Division of Radiological Sciences, Singapore General Hospital, Singapore, Singapore, 2Department of Neurology, National Neuroscience Institute, Singapore, Singapore, 3Duke-NUS Graduate Medical School, Singapore, Singapore, 4Department of Neurosurgery, National Neuroscience Institute, Singapore, Singapore, 5Department of Electrical & Computer Engineering, Seoul National University, Seoul, Republic of Korea
1Division of Radiological Sciences, Singapore General Hospital, Singapore, Singapore, 2Department of Neurology, National Neuroscience Institute, Singapore, Singapore, 3Duke-NUS Graduate Medical School, Singapore, Singapore, 4Department of Neurosurgery, National Neuroscience Institute, Singapore, Singapore, 5Department of Electrical & Computer Engineering, Seoul National University, Seoul, Republic of Korea
Synopsis
High resolution susceptibility map-weighted images (SMWI) has recently been developed to improve visualization of the nigrosome-1 region of the substantia nigra (SN) on 3T imaging. SMWI utilizes susceptibility weighting mask derived from quantitative susceptibility mapping (QSM) to enhance the contrast for the SWI magnitude images. In this study, we explore the utility of SMWI to enhance visual identification of the subthalamic nucleus (STN) at 3T, which may be useful to aid in neuro-navigational procedures requiring precise and accurate identification of the STN such as deep brain stimulation (DBS) surgery.
Introduction
High resolution susceptibility map-weighted images (SMWI) has recently been developed to improve visualization of the nigrosome-1 region of the substantia nigra (SN) on 3T imaging.1 SMWI utilizes susceptibility weighting mask derived from quantitative susceptibility mapping (QSM) to enhance the contrast for the SWI magnitude images. In this study, we explore the utility of SMWI to enhance visual identification of the subthalamic nucleus (STN) at 3T, which may be useful to aid in neuro-navigational procedures requiring precise and accurate identification of the STN such as deep brain stimulation (DBS) surgery.Methods
This feasibility study was performed on three patients with Parkinson’s Disease (PD). Written consent was obtained from all subjects. MRI was performed on a 3T (Siemens Skyra) scanner.A 3D high resolution T2* susceptibility weighted imaging was performed using a multi-echo GRE sequence with TR = 48 ms, TE = 13.77, 26.39 and 39 ms, FA = 20°, in-plane resolution = 0.5 x 0.5 x 1 mm3, number of slices = 44, scan duration = 5:42 sec. A coronal imaging plane oriented perpendicular to the anterior-posterior bi-commissural line was chosen for acquisition. For co-registration, a 3D T1-weighted MPRAGE sequence was also performed with the following parameters: TR/TE=1900/2.3ms, TI=925ms, FA=9°, in-plane resolution = 0.9 x 0.9 x 1 mm3, number of slices = 208, scan time = 4:24 sec.
QSM images were reconstructed from the multi-echo GRE images with STI Suite (University of Berkeley, California, USA).2 The magnitude images of the three echoes were combined into a single image by root sum of squares. Phase images of different TEs were unwrapped using a Laplacian unwrapping algorithm with a combined frequency calculated in each voxel. From the frequency images, the background field was removed using the harmonic background phase removal using the Laplacian operator (HARPERELLA) method.QSM was reconstructed using the improved sparse linear equation and least-squares (iLSQR) method. SMWI images were generated from the combined magnitude image and QSM image with SMWI software written in MATLAB (Laboratory for Imaging Science & Technology, Seoul National University, Seoul, South Korea).1
Results
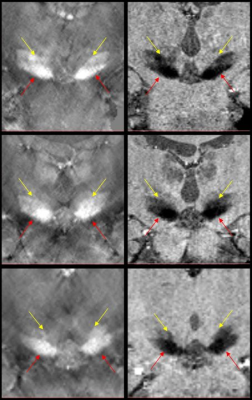
QSM and SMWI images showed clear distinction between the STN and SN (Figure 1). SMWI image of subject #2 showed enhanced visualization of the STN as compared to the QSM image (Figure 1, middle row).Dicussion
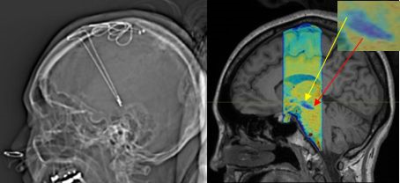
Nam et al has showed that SMWI improved the contrast-to-noise ratio between region with high iron content such as SN with its surrounding anatomical structures.1 SMWI was originally developed to improve the visualization of nigrosome-1 inside the SN but here we showed that it could also be used to enhance differentiation between SN and STN. Precise localization of the STN for DBS is important as it impacts treatment outcomes. However, the STN is small, deeply situated and difficult to be reliably or reproducibly imaged on routine clinical 1.5T and 3T scanners. Traditionally, STN targeting requires additional information from brain atlas coordinates. Subject #3 underwent DBS two weeks after MRI brain scan following traditional neurosurgical approaches, supplemented by additional pre-DBS SMWI imaging (Figure 2) with patient consent. Patient was well after the DBS procedure with good motor outcome. Figure 2 shows the final position of the DBS electrodes very closely situated to localization of the STN on the pre-DBS SMWI image.Conclusion
SMWI may offer useful supplementary visualization aid to identify the STN in surgical neuro-navigation. Further study is needed to evaluate the full potential of SMWI in aiding safe, accurate and faster DBS surgical planning.Teaching points
- SMWI enhances the contrast between SN and adjacent anatomical structures such as the STN.
- SMWI imaging plane shall be oriented perpendicular to anterior-posterior bi-commissural line to allow optimal visualisation of STN.
- Improved visualisation of STN may ease reliance on brain atlases for localisation, which have been shown to have variable positions and sizes of STN.