4876
Can radiomics and machine learning capture the unique differences between invasive lobular and invasive ductal carcinoma of the breast?1breast radiology, MSKCC, New York, NY, United States, 2breast radiology, Hospital Sirio-Libanes, Sao Paulo, Brazil, 3MSKCC, New York, NY, United States
Synopsis
In this study, we hypothesized that the specific genomic profiles of invasive lobular carcinoma (ILC) can be captured with radiomics analysis and machine learning (ML) from standardized dynamic contrast-enhanced breast MRI. Three-dimensional tumor segmentation of the first post-contrast T1-weighted sequence was conducted and included the entire mass and non-mass enhancement lesions, unifocal and multifocal/multicentric lesions. This supervised ML model produced an accuracy of 76.6%, sensitivity of 72.7%, specificity of 80.6%, PPV of 79.1% and NPV of 74.5%. Our preliminary results indicate that radiomics analysis coupled with supervised ML allows a non-invasive differentiation between ILC and invasive ductal carcinoma.
Introduction
Invasive lobular carcinoma (ILC) is the second most common histologic subtype of breast cancer: ILC differs from invasive ductal carcinoma (IDC) in its clinopathological characteristics, mestastaic patterns and responsiveness to systemic therapy. Comprehensive molecular analyses have been reported for ILCs, confirming that these tumors have specific genomic profiles compared to IDC1,2. Radiomics, the extraction and analysis of quantitative imaging features, coupled with machine learning (ML) allows imaging phenotypes to be correlated with histopathologic and genomic information3; its analysis in breast cancer has shown encouraging results for the diagnostic differentiation of malignant from benign lesions, differentiation of molecular subtypes, and other prognostic parameters, such as pathological stage and lymph node involvement4–6. We hypothesized that the specific genomic profiles of ILC can be captured with radiomics analysis and supervised ML from standardized dynamic contrast-enhanced MRI (DCE-MRI) and thereby a differentiation of invasive lobular and ductal breast cancer is feasible.Methods
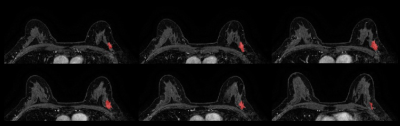
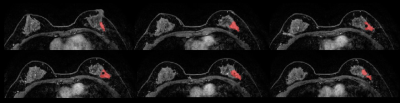
This is a retrospective Health Insurance Portability and Accountability Act (HIPAA)-compliant study approved by the Institutional Review Board for which informed consent was waived. Digital Imaging and Communications in Medicine (DICOM) images from the dynamic contrast enhanced T1 weighted MRI sequence performed were transferred to a database and loaded into the open source image processing tool OsiriX (OsiriX Foundation). 100 women with biopsy-proven ILC and 100 women with biopsy-proven IDC who underwent pretreatment breast MRI were included. Lesions with mixed histologic features were excluded. Three-dimensional tumor segmentation of the first post-contrast T1-weighted sequence was conducted by a dedicated breast radiologist with six years of experience in breast imaging to include the entire mass and non-mass enhancement lesions, unifocal and multifocal/multicentric lesions, and an adequate distance was kept from the surrounding anatomic structures and biopsy markers. One IDC case and two ILC cases were excluded due to technical issues in imaging analysis, totalizing 99 IDC and 98 ILC. Radiomics parameters were calculated using publicly available CERR software, and measures of accuracy, including sensitivity, specificity, negative predictive value (NPV) and positive predictive value (PPV), were estimated.Results
Of the 102 calculated radiomics parameters, 67 were significantly different between the two groups. After ROC curve analysis, any parameter with AUC under 0.65 was rejected and parameter reduction via correlation analysis was also employed. Finally, 12 parameters were entered into multivariate modelling. After comprehensive testing, the best model used 6 parameters and can be considered to be robust since 5-fold cross validation with a quadratic support vector machine was used in the development. The model parameters are skewness (first order parameter), lzlgle (large zone low gray level emphasis), lzhgle (large zone high gray level emphasis) and zone emphasis (size zone matrix-based parameters), and gln (gray level non-uniformity) and energy (neighborhood gray level dependence matrix-based parameters). This supervised ML model produced an accuracy of 76.6%, sensitivity of 72.7%, specificity of 80.6%, PPV of 79.1% and NPV of 74.5%.Discussion
In this study, we hypothesized that the specific genomic profiles of ILC can be captured with radiomics analysis and supervised ML from standardized DCE-MRI. Our preliminary results indicate that radiomics analysis coupled with ML allows a non-invasive differentiation of these distinct breast cancer types.The incorporation of automated feature extraction algorithms (i.e. quantitative radiomics) into routinely performed, noninvasive imaging modalities, such as DCE-MRI has the ability to stimulate the development and the use of imaging biomarkers that may provide reachable biological information without direct tissue biopsy.
Validation of the promising results of this initial study with larger patient numbers and the combination with MRI pre-contrast images might further improve the results of this radiomic analysis and is currently ongoing.
Conclusion
The developed radiomics-based ML model allows the differentiation of invasive lobular and ductal breast cancer. Further work on the radiogenomic correlations of DCE-MRI and the different specific genomic profiles of ILCs is warranted to elucidate the potential of augmented intelligence in this context.Acknowledgements
This work was partially supported by the NIH/NCI Cancer Center Support Grant (P30 CA008748) and the Breast Cancer Research Foundation.References
1. Barroso-Sousa R, Metzger-Filho O. Differences between invasive lobular and invasive ductal carcinoma of the breast: Results and therapeutic implications. Ther Adv Med Oncol. 2016;8(4):261-266.
2. Haynes B, Sarma A, Nangia-Makker P, et al. Breast cancer complexity: Implications of intratumoral heterogeneity in clinical management. Cancer Metastasis Rev. 2017 September ; 36(3): 547–555.
3. Sutton EJ, Dashevsky BZ, Oh JH, et al. Breast cancer molecular subtype classifier that incorporates MRI features. J Magn Reson Imaging. 2016;44(1):122-129.
4. Leithner D, Horvat JV, Marino MA, et al. Radiomic signatures with contrast-enhanced magnetic resonance imaging for the assessment of breast cancer receptor status and molecular subtypes: initial results. Breast Cancer Res. 2019 Sep 12;21(1):106.
5. Li H, Zhu Y, Burnside ES, et al. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. npj Breast Cancer. 2016;2(1).
6. Mazurowski MA, Zhang J, Grimm LJ, et al. Radiogenomic analysis of breast cancer: Luminal B molecular subtype is associated with enhancement dynamics at MR imaging. Radiology. 2014;273(2):365-372.