4838
Evaluating the pairwise AdaBoost model in predicting the efficacy of chemo-radiotherapy for advanced rectal cancer under small sample size
Qinglei Shi1, Xiaoming Xi*2, Yilong Yin3, Jie Kuang4, Gaofeng Shi4, Xu Yan5, Yi Qu6, and Dongsheng Zhou7
1School of Software, Shan Dong University, Jinan, China, 2School of Computer Science and Technology, Shandong Jianzhu University, Jinan, China, 3School of Software, Software Park Campus, Shandong University, Jinan, China, 4CT Room, Radiology Department, Hebei Medical University Affilited 4th Hospital, Shi Jiazhuang, China, 5MR Scientific Marketing, Siemens Healthcare, Shanghai, China, Shanghai, China, 6Department of Geriatrics, Qilu Hospital of Shandong University, Jinan, China, 7Department of Breast and Thyroid Surgery, Shandong Provinvial Qianfoshan Hospital, The First Hospital Affiliated with Shandong First Medical University, Jinan, China
1School of Software, Shan Dong University, Jinan, China, 2School of Computer Science and Technology, Shandong Jianzhu University, Jinan, China, 3School of Software, Software Park Campus, Shandong University, Jinan, China, 4CT Room, Radiology Department, Hebei Medical University Affilited 4th Hospital, Shi Jiazhuang, China, 5MR Scientific Marketing, Siemens Healthcare, Shanghai, China, Shanghai, China, 6Department of Geriatrics, Qilu Hospital of Shandong University, Jinan, China, 7Department of Breast and Thyroid Surgery, Shandong Provinvial Qianfoshan Hospital, The First Hospital Affiliated with Shandong First Medical University, Jinan, China
Synopsis
This paper proposed a pairwise AdaBoost model in predicting the therapeutic effect of non-metastatic LARC treated with neoadjuvant chemotherapy-radiation therapy based on radiomics signatures coming from ADC maps. Compared with traditional models, the pairwise AdaBoost model has ability to enlarge the number of training samples, which is useful to improve the generalization ability of the model. The experimental results demonstrated that the pairwise AdaBoost model seems can improve the accuracy and robustness of the model in predicting the treatment effect for locally LARC treated with neoadjuvant chemotherapy-radiation therapy.
Purpose
To establish and optimize a pairwise AdaBoost model, and to evaluate the value of it in predicting the therapeutic effect of non-metastatic locally advanced rectal cancer (LARC) treated with neoadjuvant chemotherapy-radiation therapy (CRT) based on radiomics signatures coming from apparent diffusion coefficient (ADC) maps.Materials and Methods
This retrospective study included 55 patients with non-metastatic LARC (male: female 32:11; age range: 28 to 77; mean age: 56.77±12.66) underwent at a 3 T scanner (MAGNETOM Skyra, Siemens Healthcare, Erlangen, Germany) from March 2013 to May 2018. According to curative effect, patients were divided into treatment effective group (TRG0 6 cases; TRG1 8 cases; TRG2 19 cases) and treatment ineffective group (TRG3 10 cases). The inclusion criteria of the study cohort were as follows: (a) MRI scan underwent within 1 week before CRT and within 1-2 weeks after CRT, and the scanned sequence included high-resolution T2WI and DWI (b-values 50 and 800 s/mm2); (b) postoperative pathological data and tumor regression level (TRG) records were complete. Radiomics signatures were extracted using an open source tool named Pyradiomics (https://pyradiomics.readthedocs.io/en/latest/index.html). In order to get samples pairs, six patients with typical effective effect (TRG0 6 cases) and three patients with typical ineffective effect were selected and regarded as template. Then, sample pair was generated by calculating the differences between each template patient’s data and the other patients’ data within and between groups. The difference intra-class was regarded as negative case, and the difference within class was regarded as positive case. Finally, the obtained 378 sample pairs were used as the training and testing data. In this study, 264 paired-case were gained as the training data set (153/111 = positive/negative)) and 111 paired-case were gained as the independent testing data set (66/48 = positive/negative). The diagnostic ability of models among different methods in normalization, dimensional reduction, and features selection were compared and optimized. AdaBoost was used as the classifier which is a meta-algorithm that conjunct other type of algorithms and combine them to get a final output of boosted classifier. AdaBoost is sensitive to the noise and the outlier. Over-fitting can also be avoided by AdaBoost. The performance of the model was evaluated using receiver operating characteristic (ROC) curve analysis. The processes were implemented with FeAture Explorer (FAE, v0.2.5, https://github.com/salan668/FAE) on Python (3.6.8, https://www.python.org/).Results
After optimization, a norm0centerunit, a pearson correlation coefficients (PCC) and an analysis of variance (ANOVA) method were used in data normalization, dimension reduction and feature selection. The model based on 9 features can get the highest AUC (0.951) and accur25acy (0.) on the validation data set. In this point, The AUC and the accuracy of the model achieved 0.997 and 0.984 on testing data set. The sensitivity and specificity were 1.000 and 0.9828 on the testing data, with 1.000 and 0.8333 for the NPV and PPV, respectively. The selected features were shown in Table 1, and the ROC curve was shown in Figure 1.Discussions
Although in the last decade big data problems attracted great interest, many real-world problems are in fact small data problems. For example, in evaluating and predicting clinical situations, collection of data sets are usually need a long time to observation and usually fewer than 100 data points. For some rare diseases, because they affect a relatively small percentage of population and in many cases, collecting data is difficult, expensive and sometimes invasive. Learning from a small data set is challenging because lack of data increases uncertainty and easily causes over fitting. Small data set is an important issue in many applications and is studied by other researchers for classification [1]. The purpose of this paper is to evaluate an effective classification model for small data sets. To deal with the problem of lack of training data, semi-supervised learning and transfer learning have been applied [3, 4]. However, small data set problems are different from these two settings. Semi-supervised learning aims to make use of unlabeled data for training, typically given a small amount of labeled data with a large amount of unlabeled data, but in small data set problems, both labeled and unlabeled data are few. Transfer learning aims to make use of other data from related domains for training. However, it is difficult to measure whether a data set is related or not and hard to guarantee no negative transfer. This paper proposed a pairwise AdaBoost model in predicting the therapeutic effect of non-metastatic LARC treated with neoadjuvant chemotherapy-radiation therapy based on radiomics signatures coming from ADC maps. Pairwise classification is the task to predict whether the examples a,b of a pair (a,b) belong to the same class or to different classes. In particular, interclass generalization problems can be addressed in this way. The reason may be that more relationship information among samples can be learned, which can enhance the generalization ability of the model [5]. The value of this model was confirmed by excellent performance on this small data set.Conclusions
The experimental results demonstrated that the pairwise AdaBoost model proposed in this paper seems can improve the accuracy and robustness of the model in predicting the treatment effect for locally LARC treated with neoadjuvant chemotherapy-radiation therapy with small sample.Acknowledgements
This study was funded by Natural Science Foundation of China (61701280)References
[1] Dougherty, Edward R., Lori A. Dalton, and Francis J. Alexander. "Small data is the problem." 2015 49th Asilomar Conference on Signals, Systems and Computers. IEEE, 2015. [2] Chang, Che-Jung, et al. "A forecasting model for small non-equigap data sets considering data weights and occurrence possibilities." Computers & Industrial Engineering 67 (2014): 139-145. [3] Srijith, P., Shevade, S., and Sundararajan, S. (2013). Semi-supervised Gaussian process ordinal regression. In Machine Learning and Knowledge Discovery in Databases, 144-159. Springer. [4] Seah, C.W., Tsang, I.W., and Ong, Y.S. (2012). Transductive ordinal regression. Neural Networks and Learning Systems, IEEE Transactions on, 23(7), 1074-1086. [5] Pairwise Support Vector Machines and their Application to Large Scale Problems. Carl Brunner, Andreas Fischer, Klaus Luig, Thorsten Thies. Journal of Machine Learning Research 13 (2012) 2279-2292.Figures
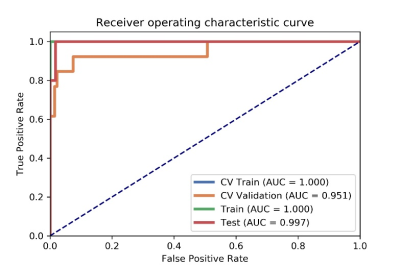
Figure
1 The AUC values of ROCs on CV training, CV validation, training and testing
data. ; CV validation: the average of k-1 folder of validation data set in
k-folder cross validation; CV training: the average of k-1 folder of training
data set in k-folder cross validation.
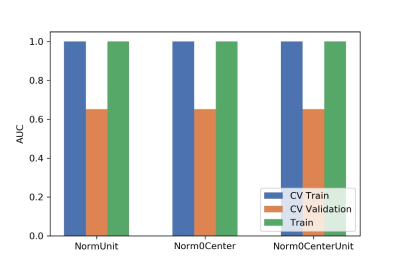
Figure
2 The effects of three feature normalization methods on CV training, CV validation data, all training and the
corresponding AUCs. CV validation: the average of k-1 folder of validation data
set in k-folder cross validation; CV training: the average of k-1 folder of
training data set in k-folder cross validation.
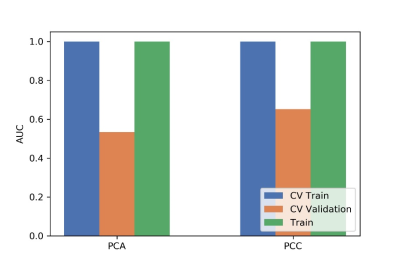
Figure
3 The effects of two dimensional reduction methods on CV training data, CV
validation data, all training data and the corresponding AUCs. CV validation:
the average of k-1 folder of validation data set in k-folder cross validation;
CV training: the average of k-1 folder of training data set in k-folder cross
validation.
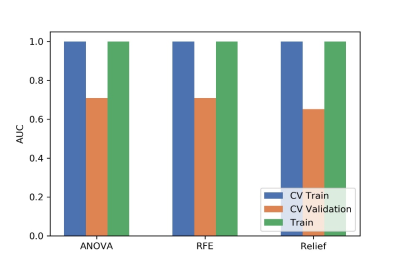
Figure
4 The effects of three feature selective methods on CV training data, CV
validation data, all training data and the corresponding AUCs. CV validation:
the average of k-1 folder of validation data set in k-folder cross validation;
CV training: the average of k-1 folder of training data set in k-folder cross
validation.
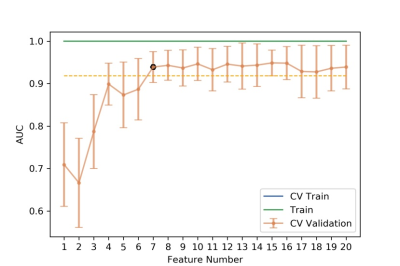
Figure 5 The number of features and concerned AUC values
according to training and testing data in the model. A minimal number of 9 has been chosen according
this diagram.
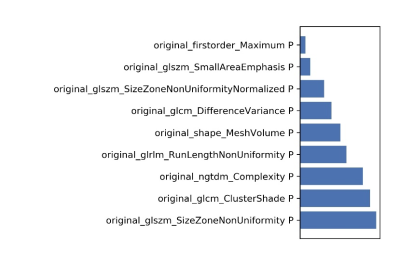
Figure 6 The selected features and their contributions on
the model.