4823
Neural-network discrimination of human plasma samples to detect pancreatic cancer
Meiyappan Solaiyappan1, Santosh K Bharti1, Mohamad Dbouk2, Paul T Winnard1, Michael Goggins2,3, and Zaver M. Bhujwalla1,3,4
1The Russell H. Morgan Dept of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2Departments of Pathology and Medicine, The Sol Goldman Pancreatic Cancer Research Center, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 3Sidney Kimmel Comprehensive Cancer Center, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 43Radiation Oncology and Molecular Radiation Sciences, The Johns Hopkins University School of Medicine, Baltimore, MD, United States
1The Russell H. Morgan Dept of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2Departments of Pathology and Medicine, The Sol Goldman Pancreatic Cancer Research Center, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 3Sidney Kimmel Comprehensive Cancer Center, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 43Radiation Oncology and Molecular Radiation Sciences, The Johns Hopkins University School of Medicine, Baltimore, MD, United States
Synopsis
The insidious growth of pancreatic cancer is a major factor contributing to its lethality. Only 10-15% of pancreatic cancers are resectable by the time they are detected. Early detection of pancreatic cancer through routine screening is clearly an unmet clinical need. Here we have applied neural network analysis to 1H magnetic resonance spectra of human plasma samples to differentiate between healthy subjects (control), subjects with benign lesions, and subjects with pancreatic ductal adenocarcinoma (PDAC). Our data support developing a neural-network approach to identify PDAC from 1H MRS of plasma samples.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the most frequent form of pancreatic cancer and its low survival rate of less than 4% at five years makes it the fourth leading cause of cancer-related deaths. The poor prognosis of PDAC is mainly due to late-stage diagnosis [1, 2]. Similarities in the clinical behavior and imaging features of PDAC and chronic pancreatitis further complicate the detection of PDAC [3]. Although inroads are being made in developing molecular imaging probes, these have not been clinically translated. There is an urgent need for noninvasive clinically translatable biomarkers of PDAC. Plasma based tests provide an attractive option for routine screening. Here we are evaluating the application of neural-network analysis to 1H MR spectra of human plasma samples to identify PDAC that could potentially be developed for screening, initially in high risk patients.Methods
Plasma samples from healthy subjects (control, n=18), from subjects with benign pancreatic lesions (benign, n=22), and from subjects with PDAC (PDAC/malignant, n=24) were analyzed with 1H MRS. 1H MR spectra were acquired on a Bruker Avance III 750 MHz (17.6 T) MR spectrometer equipped with a 5 mm probe. For NMR analysis, 250 uL of plasma was mixed with 350uL of D2O phosphate buffered saline. Spectra with water suppression were achieved using pre-saturation and were acquired using a single pulse sequence and Carr-Purcell-Meiboom-Gill (CPMG) pulse sequence with the following experimental parameters: spectral width of 15495.86 Hz, data points of 64 K, 90o flip angle, relaxation delay of 10 sec, acquisition time 2.11 sec, 64 scans with 8 dummy scans, receiver gain 64. All spectral acquisition, processing and quantification were performed using TOPSPIN 3.5 software. Area under peaks were integrated and normalized with respect to TSP as well as to the tissue weights used for dual phase extraction. CPMG NMR spectra were used for analysis and representative spectra from the three groups are presented in Figure 1. After the initial processing of the spectral data to calibrate against reference peak signal and the plasma volume quantity, various spectral features were extracted to understand if a suitable technique can be developed to discriminate the three classes of spectra. Among these, three spectral features showed high probability for a possible successful discrimination. These spectral features are: (a) signal intensity weighted-mean of the spectra, (b) standard deviation of the spectra about the weighted-mean from (a), and (c) weighted mean of the difference spectra, i.e., the absolute difference of the spectra after subtraction from mean control spectra. Figure 2(a) shows the 3D scatter plot of these three spectral features (in arbitrary spectral units). We observed that the distribution pattern exhibited a tendency toward discrimination that can be further accentuated when more number of cases are added, thus suggesting the discrimination problem can be more effectively solved by an Artificial Intelligence based technique that can learn from the data. This lead to the design of our three-layer artificial neural network model reported here to solve the discrimination problem. The artificial neural-network, developed in MATLAB 2019b (MathWorks, Inc), was limited to a stack of two auto-encoder layers and a ‘softmax’ layer. Due to the limited size of the data all of the data were used to train the network while keeping low L2-regularization to avoid over-training. Cross-validation results were captured as confusion matrices and receiver operating characteristic (ROC) curves.Results and Discussion
The receiver operating characteristics (ROC) curves in Figure 2(b) and the confusion plot in Figure 3 demonstrate the performance of the neural-network based discrimination. It should be noted that our training data have different samples sizes and this can impact the prediction accuracy of respective classes. Thus in fine-tuning the network, we aimed for overall balanced performance, particularly between control cases and the other two since its sample size was lesser than other two.We have demonstrated that a combination of spectral features extraction and neural network processing of MRS data of plasma samples can make it feasible to successfully discriminate between control, benign and malignant pancreatic cancer. While the limited number of cases in our sample size is a concern, the fact that this approach yielded satisfactory results suggests that the approach can be extended fairly readily to encompass a large sample size to both improve the accuracy and as well as provide a robust solution for plasma based prediction of the presence of PDAC.
Acknowledgements
This work was supported by NIH R35CA209960 and R01CA193365.References
1. Vincent A, Herman J, Schulick R, Hruban RH, Goggins M. Pancreatic cancer. Lancet. 2011;378(9791):607-20. Epub 2011/05/31. doi: 10.1016/S0140-6736(10)62307-0. PubMed PMID: 21620466. 2. Whatcott CJ, Posner RG, Von Hoff DD, Han H. Desmoplasia and chemoresistance in pancreatic cancer. In: Grippo P, Munshi H, editors. Pancreatic Cancer and Tumor Microenvironemnt2012. 3. Steer ML, Waxman I, Freedman S. Chronic pancreatitis. N Engl J Med. 1995;332(22):1482-90. Epub 1995/06/01. doi: 10.1056/NEJM199506013322206. PubMed PMID: 7739686.Figures
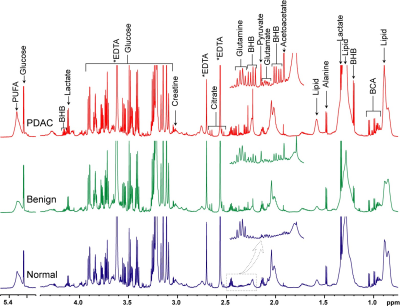
Figure 1:
Representative 1H MRS spectra obtained from plasma of healthy
subjects (control/normal), patients with benign disease and PDAC patients. Expansion of the spectra from 2.2 ppm to
2.5ppm are 4X vertically zoomed highlighting changes in metabolic patterns.
(BHB; betahydroxybutyrate, BCA; branch chain amino acid, PUFA; Polyunsaturated
fatty acid).
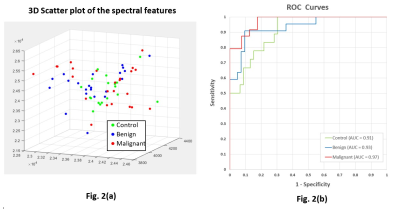
Figure 2. (a) 3D scatter plot shows three major
spectral features derived from MRS data of plasma. A significant number of control samples (green)
cluster around the central region, the malignant cases (red) are more
distributed, and the benign cases (blue) overlap both. Neural-network training teases out the
overlapping distribution to provide a satisfactory discrimination. (b)
Receiver Operating Characteristics (ROC) curves show the sensitivity and
specificity of the neural-network, with the area under the curve (AUC) for all
three classifications above 0.90.
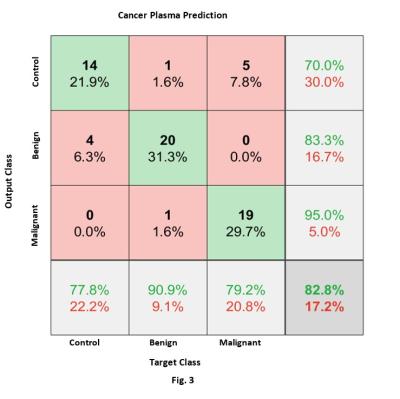
Figure 3: Confusion
Matrix result of cancer plasma prediction. The green diagonal boxes show the correct
predictions in each class and red boxes indicate misclassifications. The numbers in each box correspond to the
number of samples (and their percentage of the total data). The bottom-row shows prediction accuracy for
each class and the bottom-right corner box gives the overall correct and
incorrect predictions. Cancer Plasma
classification resulted in an 83% correct prediction.