4727
Ubiquitous Mitochondrial Creatine Kinase Drives Malignant Creatine Metabolite Profiles in Triple-Negative Breast Cancer Models1he Russell H. Morgan Department of Radiology and Radiological Science, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 2F.M. Kirby Research Center for Functional Brain Imaging, Kennedy Krieger Institute, The Johns Hopkins University School of Medicine, Baltimore, MD, United States, 3The Sidney Kimmel Comprehensive Cancer Center, The Johns Hopkins University School of Medicine, Baltimore, MD, United States
Synopsis
Ubiquitous Mitochondrial Creatine Kinase (CKMT1) is a mitochondrial membrane protein that catalyzes the reversible conversion of creatine (Cr) to phosphocreatine (PCr). This study shows in a mouse model of triple-negative breast cancer that CKMT1-overexpression significantly increases tumor Cr and PCr levels while accelerating tumor growth. Since high CKMT1 predicts worsened survival in breast cancer patients, CKMT1 may hold promise as a potential diagnostic marker and/or treatment target whose expression can be monitored using magnetic resonance spectroscopy-detected Cr and PCr levels.
Purpose
Breast cancers display altered creatine metabolism, including low levels of creatine (Cr)1 and phosphocreatine (PCr)1,2. The molecular drivers of malignant creatine metabolite profiles, and the overall role of creatine metabolism in oncogenesis and tumor progression are not well characterized. Previous research has shown a strong positive correlation between levels of creatine metabolites and mRNA expression of ubiquitous mitochondrial creatine kinase (CKMT1)3, which catalyzes the reversible interconversion of Cr to PCr4. In this study, we have employed high-resolution (HR) 1H magnetic resonance spectroscopy (MRS) to measure changes in levels of Cr and PCr in mouse breast tumor xenograft models of CKMT1 overexpression.Methods
Two triple-negative breast cancer cell lines (MDA-MB-231 and SUM159) were transduced with the pLX304 lentiviral vector carrying the CKMT1 gene under control of the CMV promoter to overexpress CKMT1. Transduction with empty pLX304 lentiviral vector was performed to establish empty-vector controls for both cell lines. Immunoblotting of CKMT1-immunoreactive bands and SYBR Green quantitative RT-PCR were used to confirm CKMT1 overexpression relative to cells transfected with empty-vector.Next, 2 × 106 breast cancer cells were orthotopically implanted into the fourth right mammary fat pad of six-week-old female athymic nude mice (CKMT1-overexpressing MDA-MB-231, n=6; empty vector-transfected MDA-MB-231, n=6). Primary tumor volumes were measured twice weekly using the formula π ((lwh))/6, where l, w, and h correspond to three orthogonal tumor dimensions.
After 8 weeks, mice were sacrificed and primary tumors were excised. Metabolites were extracted from primary tumors using dual-phase extraction (methanol:chloroform: water = 1:1:1). HR 1H MRS of the aqueous extract fractions was measured on Bruker 750 MR spectrometer, and Cr and PCr concentrations were quantified using TopSpin software.
The relationship of CKMT1 gene expression and relapse-free survival was evaluated in a multi-study breast cancer transcriptomic data set using Kmplotter5 (http://kmplot.com). Kaplan–Meier estimates of relapse-free survivals (RFS) were calculated in untreated patients (n=1010).
Results
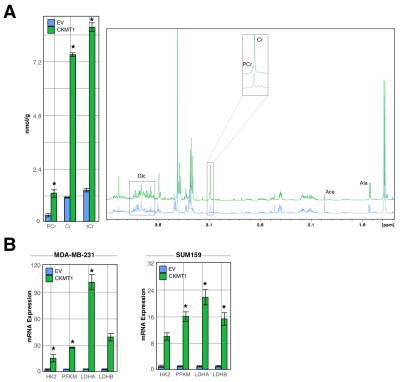
HR 1H MRS revealed significant increases in tumor levels of Cr and PCr in mice orthotopically growing MDA-MB-231 tumor xenografts overexpressing CKMT1 as compared to those growing empty vector-transfected MDA-MB-231 xenografts. Additionally, we identified increases in metabolites derived from glycolytic substrates, including alanine; but decreases in those associated with oxidative energy metabolism, such as acetate (Fig. 1A).These changes were consistent with in vitro findings in CKMT1-overexpressing MDA-MB-231 and SUM159 cells in culture that included increases in mRNA expression of genes associated with glycolysis, including hexokinase 2 (HK2), phosphofructokinase (PFKM) and lactate dehydrogenase A (LDHA) and B (LDHB) (Fig. 1B).
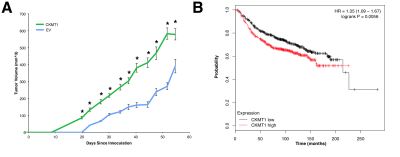
In addition, we observed an increase in tumor growth in mice bearing CKMT1-overexpressing tumors as compared to control tumors (Fig. 2A). In line with CKMT1’s prognostic significance, we noted that elevated CKMT1 expression in patients predicts worsened RFS (Fig. 2B).
Discussion
Consistent with previous studies, which identified a positive correlation between CKMT1 expression and Cr metabolite levels in vitro6, our study in orthotopic mouse tumor xenograft models of CKMT1 overexpression demonstrates in vivo that genetically-engineered CKMT1 overexpression drives increased levels of Cr and PCr. Moreover, CKMT1 overexpression conferred increased expression of enzymes involved in glycolysis in vitro and increased levels of glycolysis-associated metabolites in vivo. These changes occurred alongside increases in tumor growth, consistent with worsened patient outcomes predicted by increased levels of CKMT1 expression. Since CKMT1 may be of prognostic significance, it is possible that Cr metabolites can, in the future, be used to forecast outcomes and stratify patients by serving as a surrogate marker of CKMT1 expression.Conclusions
Elevated CKMT1 expression increases tumor growth in vivo in tumor xenograft models in mice and predicts reduced survival in patients. Since its overexpression increases tumor levels of Cr metabolites, CKMT1 is an interesting marker or target that may be monitored using MRS.Acknowledgements
We thank all members of the Division of Cancer Imaging Research in The Russell H. Morgan Department of Radiology and Radiological Science for their help and support.References
1Cao, M. D., Lamichhane, S., Lundgren, S., Bofin, A., Fjøsne, H., Giseødegård, G. F., Bathen, T. F. (2014). Metabolic Characterization of Triple Negative Breast Cancer. BMC Cancer. 14(941)
2Chan, K. W. Y., Jiang, L., Cheng, M., Wijnen, J. P., Liu, G., Huang, P., van Zijl, P. C. M., McMahon, M. T., Glunde, K. (2016) CEST-MRI Detects Metabolite Levels Altered by Breast Cancer Cell Aggressiveness and Chemotherapy Response. NMR Biomed., 10(29): 806-816.
3Ayyappan, V., Cheng, M., Cai, R., Tressler, C., Sonkar, K., McMahon, M.T., Glunde, K. (2018). “Ubiquitous Mitochondrial Creatine Kinase (CKMT1) Expression Correlates with MRS-Detectable Creatine Metabolic Profiles in Human Breast Cancer.” Proceedings of the 2018 Joint Annual Meeting ISMRM-EMRB.
4Wyss, M., Kaddurah-Daouk, R. (2000). Creatine and Creatinine Metabolism. Physiological Review, 80(3):1107-1213.
5Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2009; 123: 725–731.
6Ayyappan, V., Cheng, M., Cai, R., Tressler, C., Sonkar, K., McMahon, M.T., Glunde, K. (2019). “Ubiquitous Mitochondrial Creatine Kinase (CKMT1) Drives Malignant Creatine Metabolite Profiles in Human Breast Cancer Cell Lines.” Proceedings of the 2018 Joint Annual Meeting ISMRM-EMRB.
Figures